Data Visualization with ggplot2
ODSC West: https://bit.ly/odscw-ggp2
Outline
Part 1
Why do we need graphs?
An exploratory mindset
Surprise or confirm, then communicate
The grammar of graphics
- Part 2
- RStudio Cloud
- Exercises & solutions
- Creating a graph (layer-by-layer)
- Applying the grammar
Part 1
- Intros 👋
- Workshop materials ⬇️
- Basic understand of
ggplot2syntax ✔️ - Build your first graph! ✔️
Why do we need graphs?
Raw data don’t communicate well
It’s hard to make sense of millions of rows and/or thousands of columns
Fortunately, we are excellent at seeing patterns:
“the human brain has a superior ability to mentally manipulate animate and inanimate patterns into a myriad of intangible symbols that can then be recombined to produce new images of the world;”
“we therefore live partly in worlds of our own mental creation, super-imposed upon or distinct from the natural world.”
Graphs allow us to explore complexity with symbols and images
Exploratory Data Analysis
“Exploratory Data Analysis (EDA)” first coined by American mathematician John Tukey in 1977
“The greatest value of a picture is when it forces us to notice what we never expected to see.”
- John Tukey, 1977
Exploration requires ‘listening’
“The role of the data analyst is to listen to the data in as many ways as possible until a plausible ‘story’ of the data is apparent”
- John T. Behrens, Principles and Procedures of Exploratory Data Analysis
Exploration is a ‘state of mind’
“More than anything, EDA is a state of mind. During the initial phases of EDA you should feel free to investigate every idea that occurs to you. Some of these ideas will pan out, and some will be dead ends…”
“As your exploration continues, you will hone in on a few particularly productive areas that you’ll eventually write up and communicate to others.”
- Hadley Wickham, R for Data Science
An Exploratory Mindset
Exploration requires a Bayesian Mindset (1 of 3)
We all have implicit beliefs, or priors, about the world
What we think we know (i.e.,our expectations)

Exploration requires a Bayesian Mindset (2 of 3)
When we encounter new information or data, our priors get updated
Our expectations + new data (i.e., what we see)

Exploration requires a Bayesian Mindset (3 of 3)
Our updated beliefs, or posteriors, depend on our priors and our perceptions of the new information
What we expect + what we see = what we’ve learned

Graphs can confirm our expectations
What if our expectation was that X is related to Y?
…then we graphed the data…
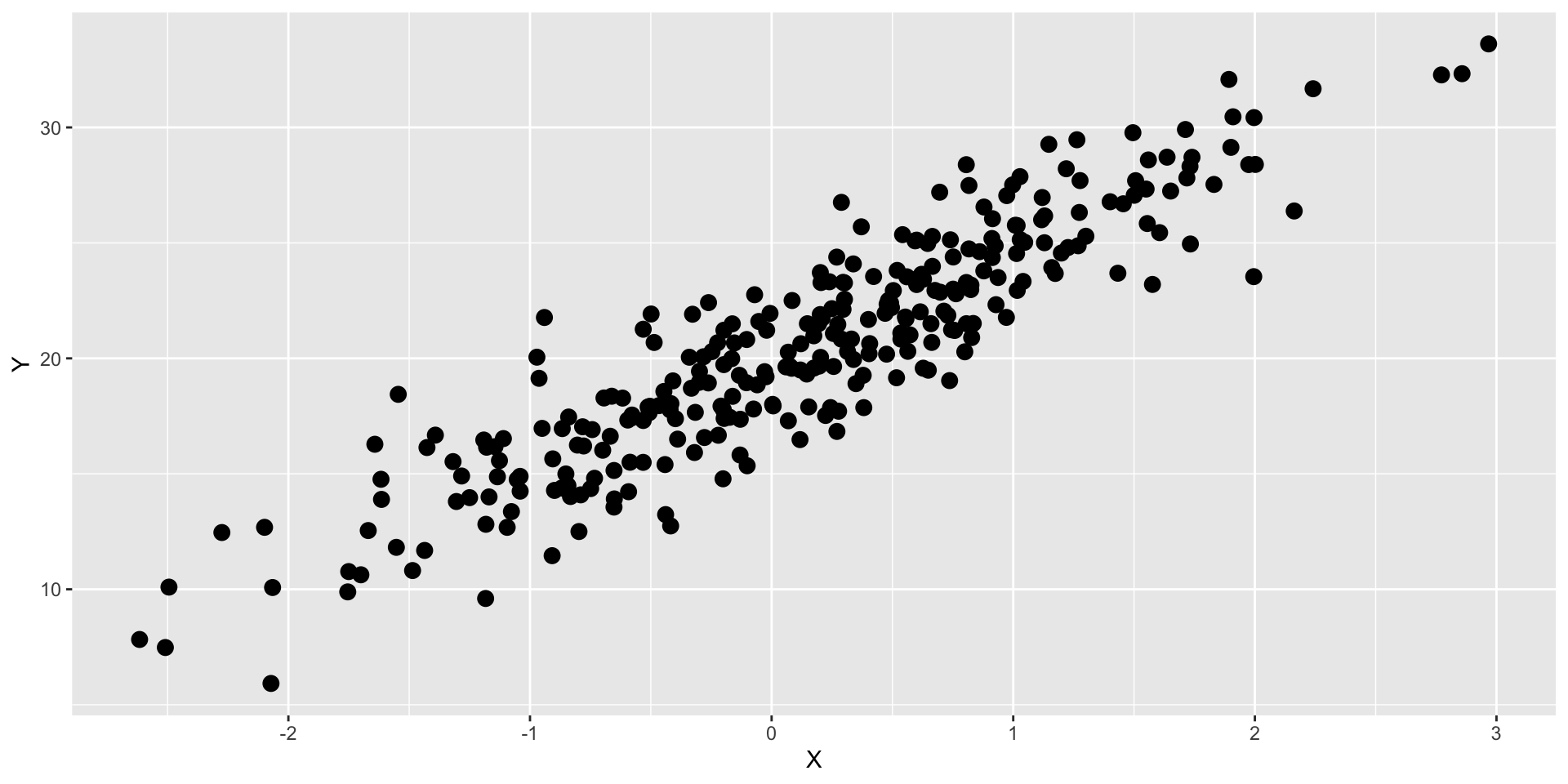
We would say our expectations have been confirmed
Graphs can refute our expectations
What if our expectation was that X is related to Y?
…then we graphed the data…
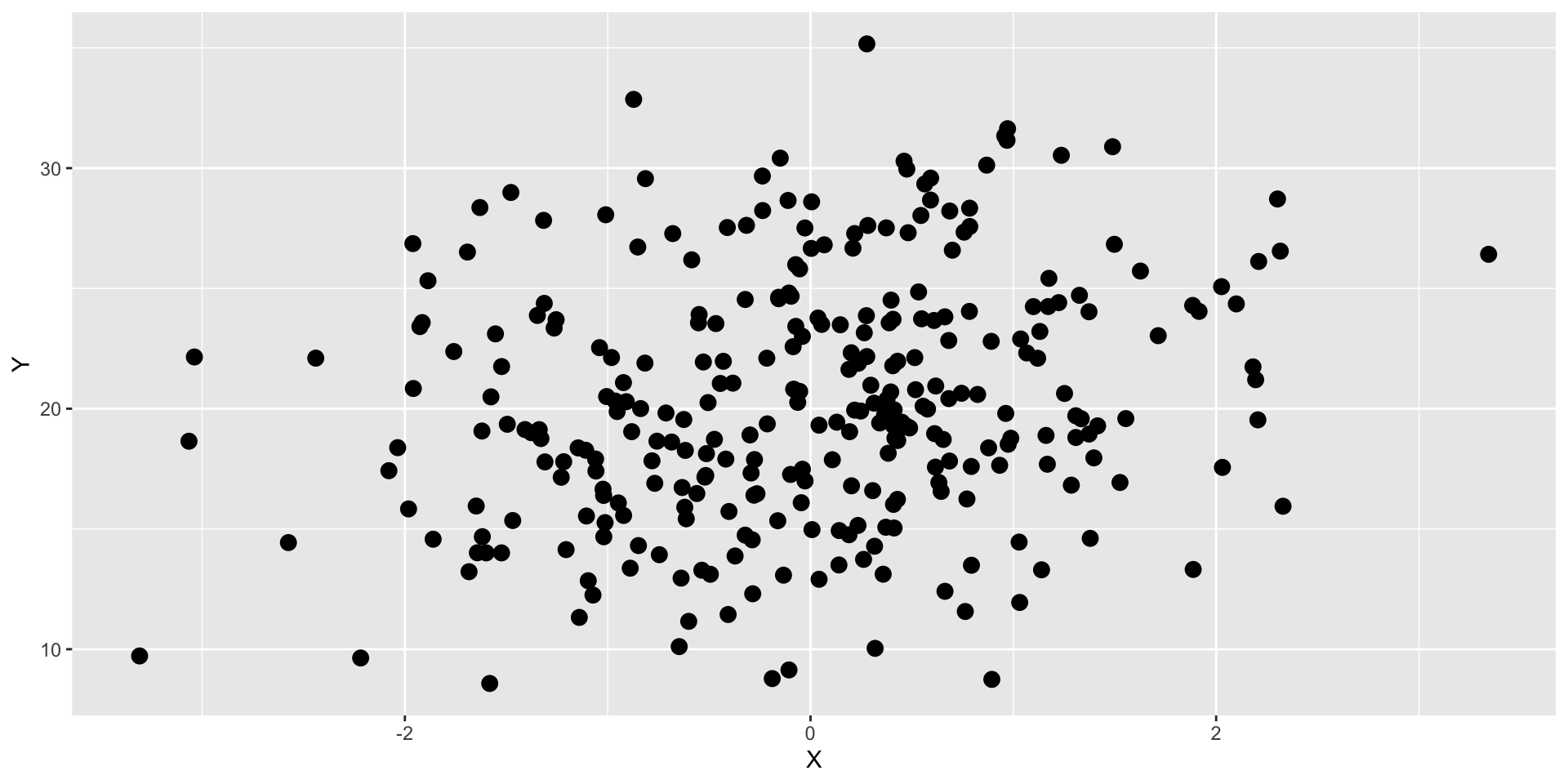
We would say our expectations have been refuted
ggplot2: grammar & syntax
Grammar
The system of rules for any given language
Includes:
- Word meanings
- Internal structure
- Word arrangement
Syntax
The form, structure and order for constructing statements

[[students][[cook][and][serve grandparents]]]
[[students][[cook and serve][grandparents]]]
ggplot2 : grammar & syntax
Built on top of the grammar & syntax of R
“In R, objects are like nouns, and functions (fn) are like verbs”
functions do things to objects
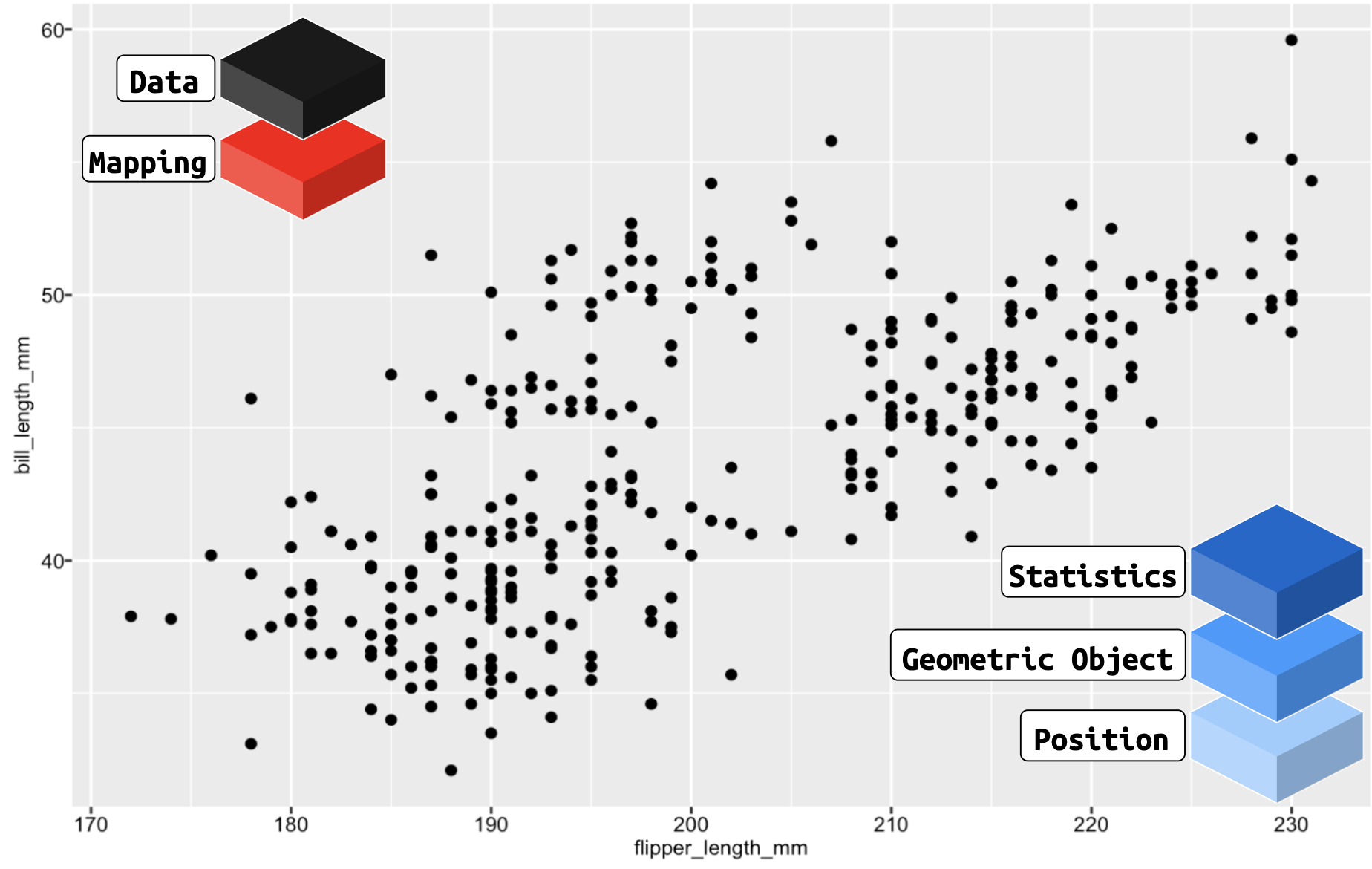
ggplot2: a layered language for graphs
ggplot2 is comprised of layers
- Data
- Mapping
- Statistics
- Geometric objects
- Position adjustments
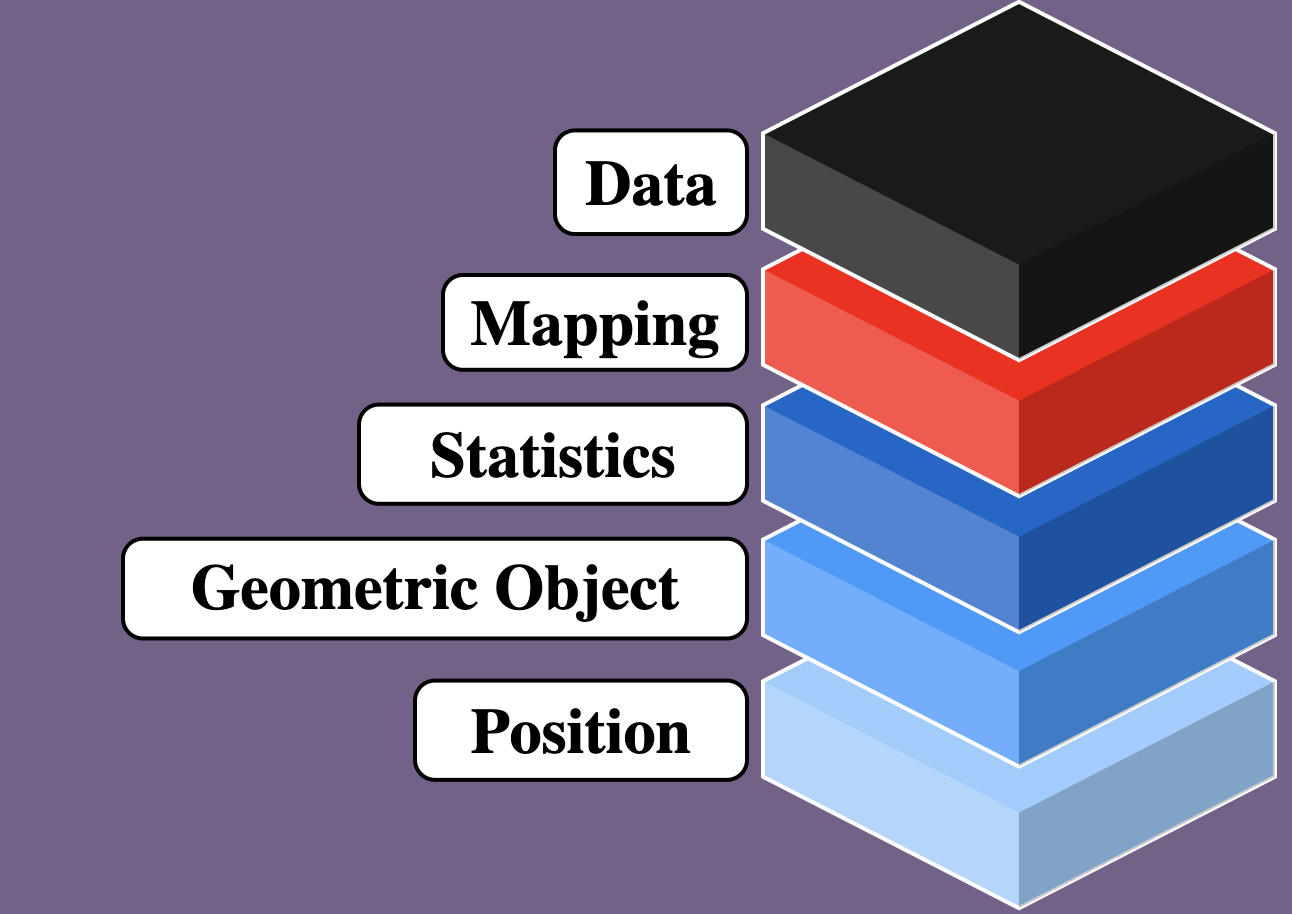
ggplot2: data
ggplot2: mapping
ggplot2: geoms
ggplot2: layers
Layers = infinitely extensible
ggplot2 is a system for,
“making infinite use of finite means” - Wilhelm von Humboldt
With a finite number of objects & functions, we can combine ggplot2’s grammar and syntax to create an infinite number of graphs!
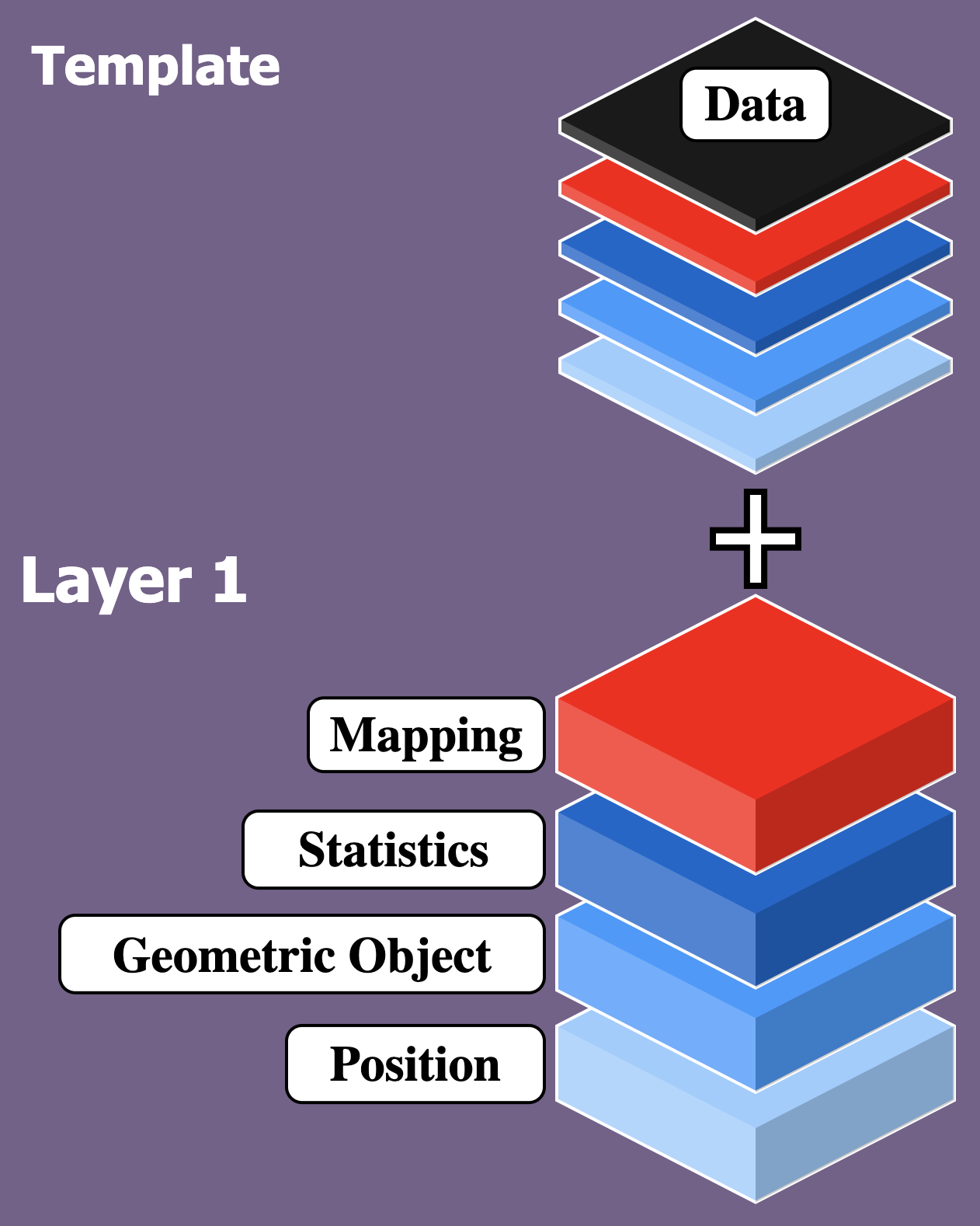
ggplot2: templates
Basic Template: Data, aesthetic mappings, geom
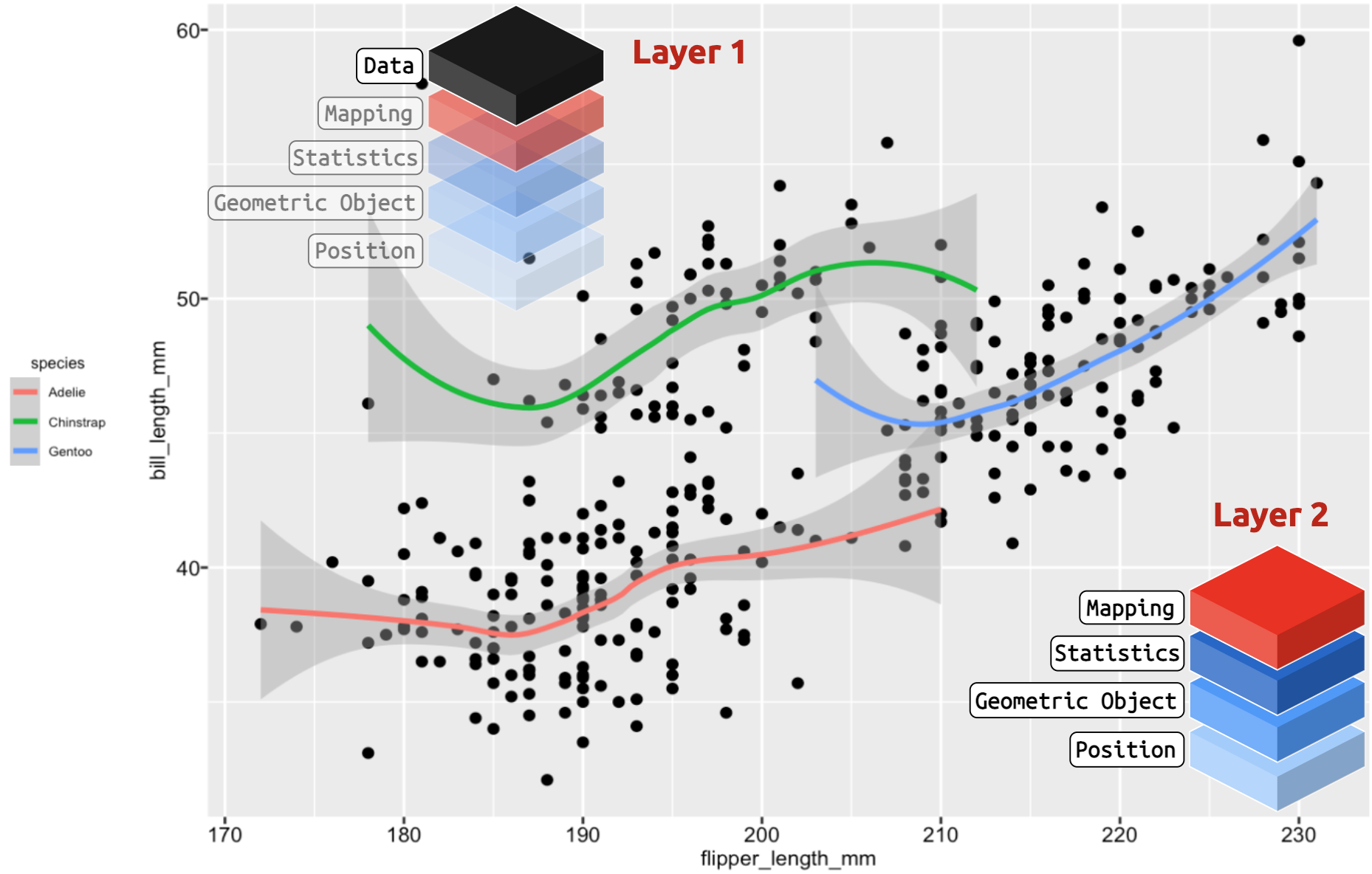
ggplot2: more templates
Template + 1 Layer: more geoms and more aesthetic mappings
ggplot2: even more!
Template + 1 Layer + Facet Layer: template, more aesthetic mappings, and facets!
Templates = infinitely extensible!
Themes
Part 2
- RStudio Cloud ✔️
- Exercises & solutions ✔️
- Creating a graph (layer-by-layer) ✔️
- Applying the grammar ✔️
RStudio.Cloud
RStudio.Cloud: Set up (1 of 4)
Head to RStudio.Cloud, you will see the following:

Log in with your GitHub credentials
RStudio.Cloud: Set up (2 of 4)
On the top of the RStudio IDE, you will see the following:

Click on Save a Permanent Copy to add this project to your workspace
RStudio.Cloud: Set up (3 of 4)
In the Files pane, click on the inst.R file to open it

RStudio.Cloud: Set up (4 of 4)
In the Source pane, click on the Source icon to run inst.R

This sends the code in inst.R to the Console
RStudio.Cloud: Exercises
The exercises are in the exercises/ folder

RStudio.Cloud: Solutions
Each exercise has a solution file in solutions/ folder

Quick Tip
Tip: writing code can be frustrating, especially in the beginning…
…it doesn’t always produce a tangible result…
…but creating visualizations is rewarding!!!
ggplot2: build the labels first!
Create a title, subtitle (with data source), and x/y axis labels
ggplot2: build graph, check labels
Build labels, build graphs, then check labels!
What’s wrong here?
ggplot2: build graph, check labels, revise
x and y are flipped!
Fixed!
On the importance of revision:
Revision Sharpens Thinking:
“More particularly, rewriting is the key to improved thinking. It demands a real open-mindedness and objectivity.”
“It demands a willingness to cull verbiage so that ideas stand out clearly. And it demands a willingness to meet logical contradictions head on and trace them to the premises that have created them.”
“In short, it forces a writer to get up his courage and expose his thinking process to his own intelligence.”
The data
Viewing data (1 of 3)
View() opens the RStudio data viewer

Viewing data (2 of 3)
glimpse() and str() are displayed in the console
Rows: 344
Columns: 8
$ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Adel…
$ island <fct> Torgersen, Torgersen, Torgersen, Torgersen, Torgerse…
$ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9, 39.2, 34.1, …
$ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8, 19.6, 18.1, …
$ flipper_length_mm <int> 181, 186, 195, NA, 193, 190, 181, 195, 193, 190, 186…
$ body_mass_g <int> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 3475, …
$ sex <fct> male, female, female, NA, female, male, female, male…
$ year <int> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007…Viewing data (3 of 3)
glimpse() and str() are displayed in the console
tibble [344 × 8] (S3: tbl_df/tbl/data.frame)
$ species : Factor w/ 3 levels "Adelie","Chinstrap",..: 1 1 1 1 1 1 1 1 1 1 ...
$ island : Factor w/ 3 levels "Biscoe","Dream",..: 3 3 3 3 3 3 3 3 3 3 ...
$ bill_length_mm : num [1:344] 39.1 39.5 40.3 NA 36.7 39.3 38.9 39.2 34.1 42 ...
$ bill_depth_mm : num [1:344] 18.7 17.4 18 NA 19.3 20.6 17.8 19.6 18.1 20.2 ...
$ flipper_length_mm: int [1:344] 181 186 195 NA 193 190 181 195 193 190 ...
$ body_mass_g : int [1:344] 3750 3800 3250 NA 3450 3650 3625 4675 3475 4250 ...
$ sex : Factor w/ 2 levels "female","male": 2 1 1 NA 1 2 1 2 NA NA ...
$ year : int [1:344] 2007 2007 2007 2007 2007 2007 2007 2007 2007 2007 ...Build from scratch, layer-by-layer
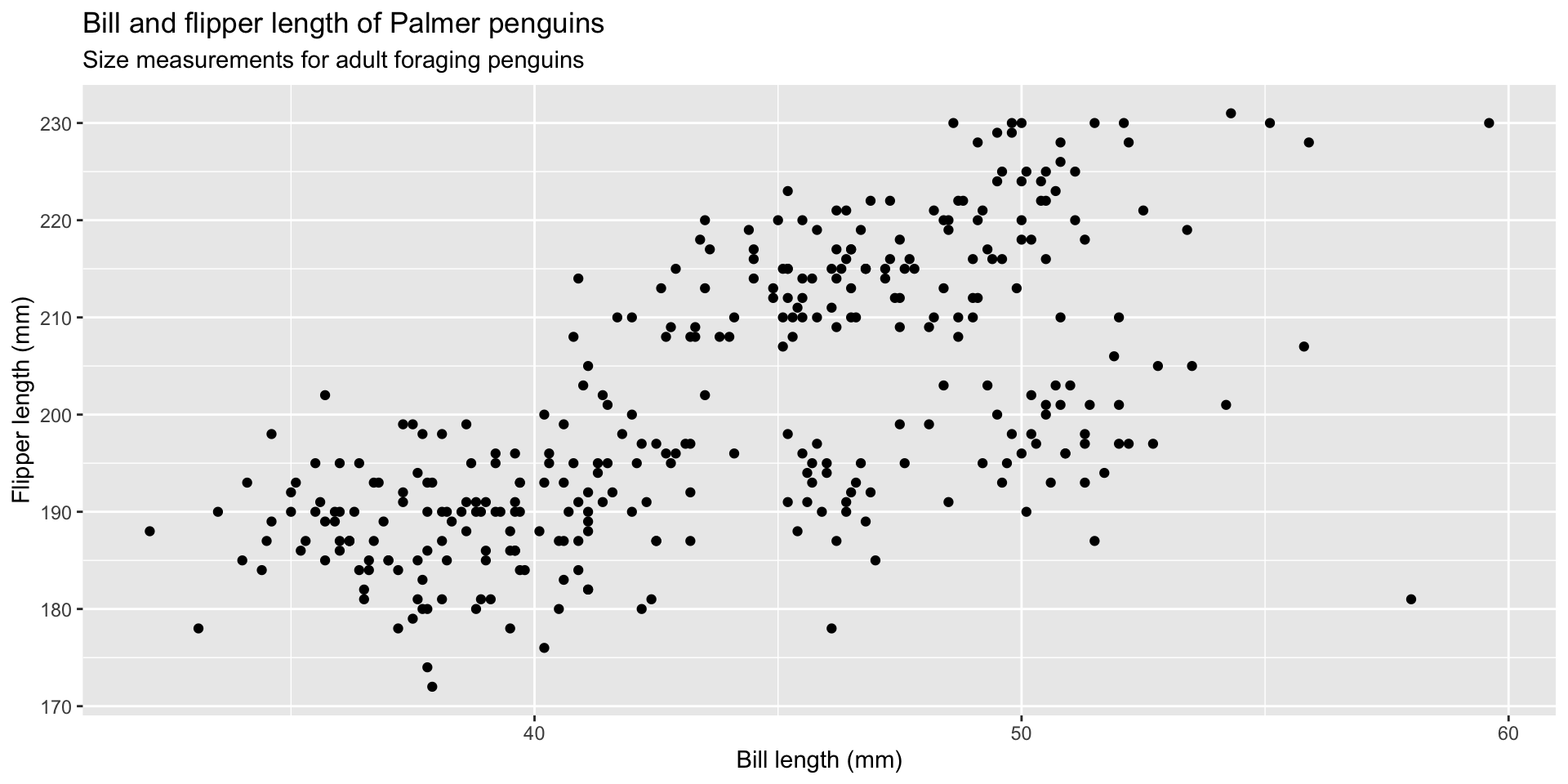
graph 01: LABELS!
We want to build the labels first:
- title = “Bill and flipper length of Palmer penguins”
- subtitle = “Size measurements for adult foraging penguins”
- x = “Bill length (mm)”
- y = “Flipper length (mm)”
graph 01: Initialize plot with data
graph 02: Map variables to positions
graph 03: Adding geoms
graph 04: Don’t forget the labels!
Global vs. local mapping
Global mapping
The previous graphs mapped aesthetics globally
Local mapping
Mapping aesthetics globally and then adding the geom_*() function results in the same graph as when we map aesthetics locally (inside the geom_*() function)
The ggplot2 templates (refresher)
The template from part 1 uses local mappings (i.e. aesthetic mappings are set inside the geom_* function).
Below we’ve adjusted the template to include global mappings (and the option to include aesthetic mappings locally)
Read more here.
graph 05: Convert global to local mappings
For graph-05.R, convert the global aesthetics to local aesthetics inside the geom_point() function
Visual encodings
What are visual encodings?
Visual encodings are what we see on the graph
Things like position, size, shape, color, etc.

Ranked by accuracy
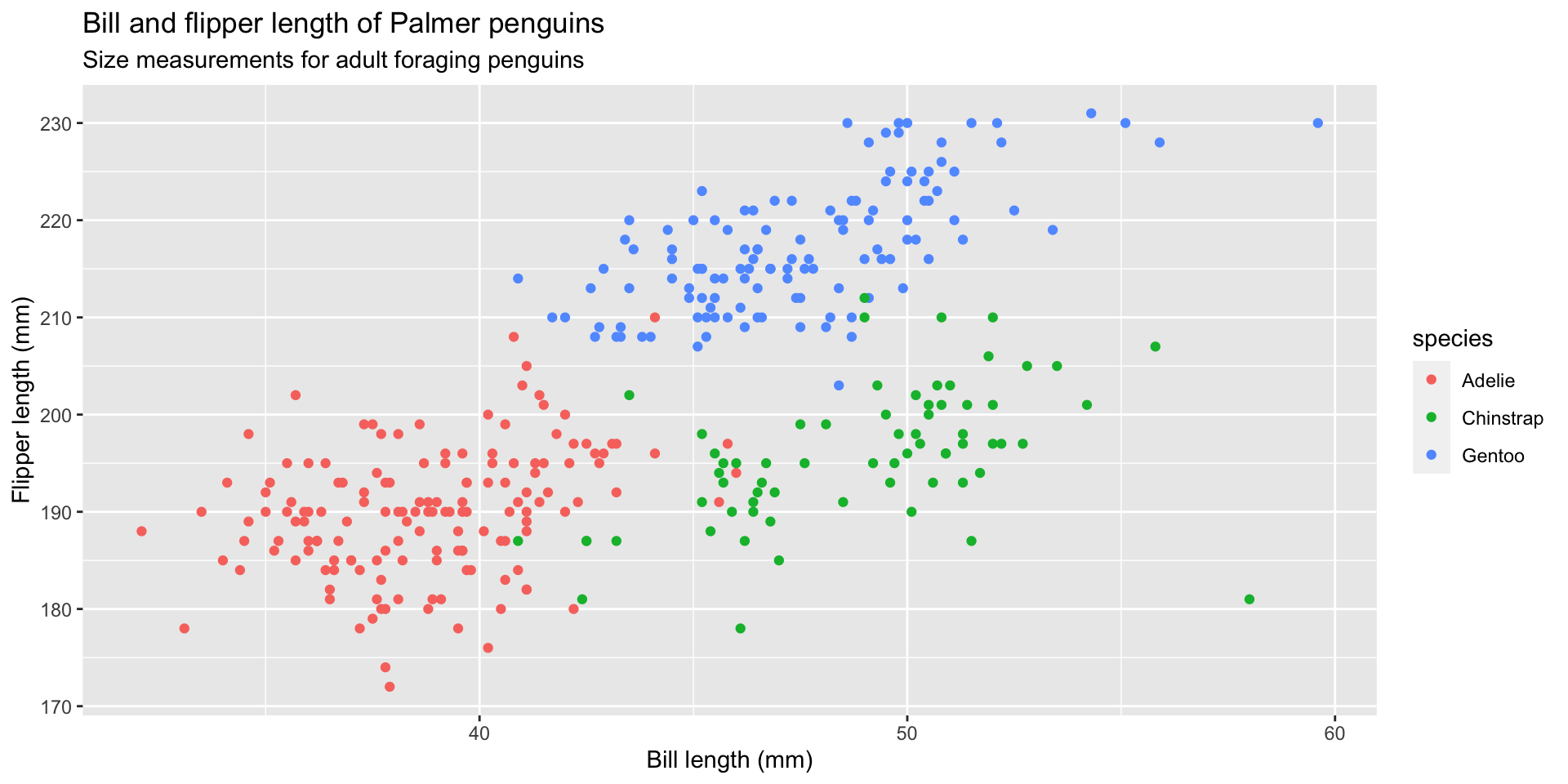
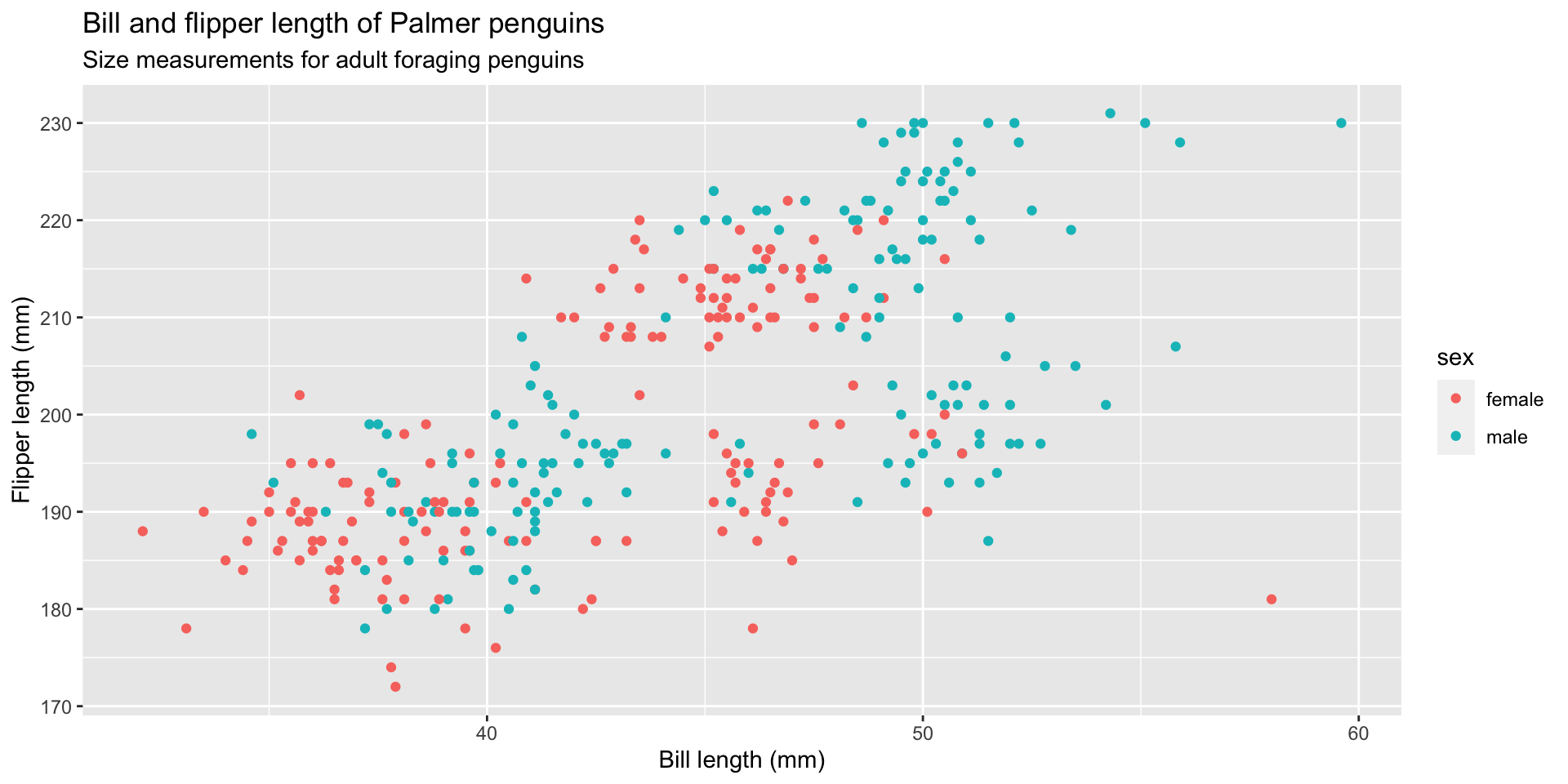
graph 06: Adding color (global)
graph 07: Adding color (local)
Map color to the species variable using local aesthetic mapping
graph 08: Color vs. Fill (1 of 2)
Below we’ll look at the counts of sex vs. species of Palmer penguins
View our data:
Rows: 333
Columns: 8
$ species <fct> Adelie, Adelie, Adelie…
$ island <fct> Torgersen, Torgersen, …
$ bill_length_mm <dbl> 39.1, 39.5, 40.3, 36.7…
$ bill_depth_mm <dbl> 18.7, 17.4, 18.0, 19.3…
$ flipper_length_mm <int> 181, 186, 195, 193, 19…
$ body_mass_g <int> 3750, 3800, 3250, 3450…
$ sex <fct> male, female, female, …
$ year <int> 2007, 2007, 2007, 2007…graph 08: Color vs. Fill (2 of 2)
graph 09: Bar position
Stacked bar-graphs make it difficult to do side-by-side comparisons using the y axis
graph 10: Histograms (special bar-graphs)
The geom_histogram() function uses ‘bins’ to represent counts for each value
Create new labels

graph 11: Density plots
Mapping vs. setting aesthetics
Mapping vs. setting (1 of 2)
Mapping vs. setting (2 of 2)
From ggplot2 book
“If you want appearance to be governed by a variable, put the specification inside
aes(); if you want override the default size or colour, put the value outside ofaes().”
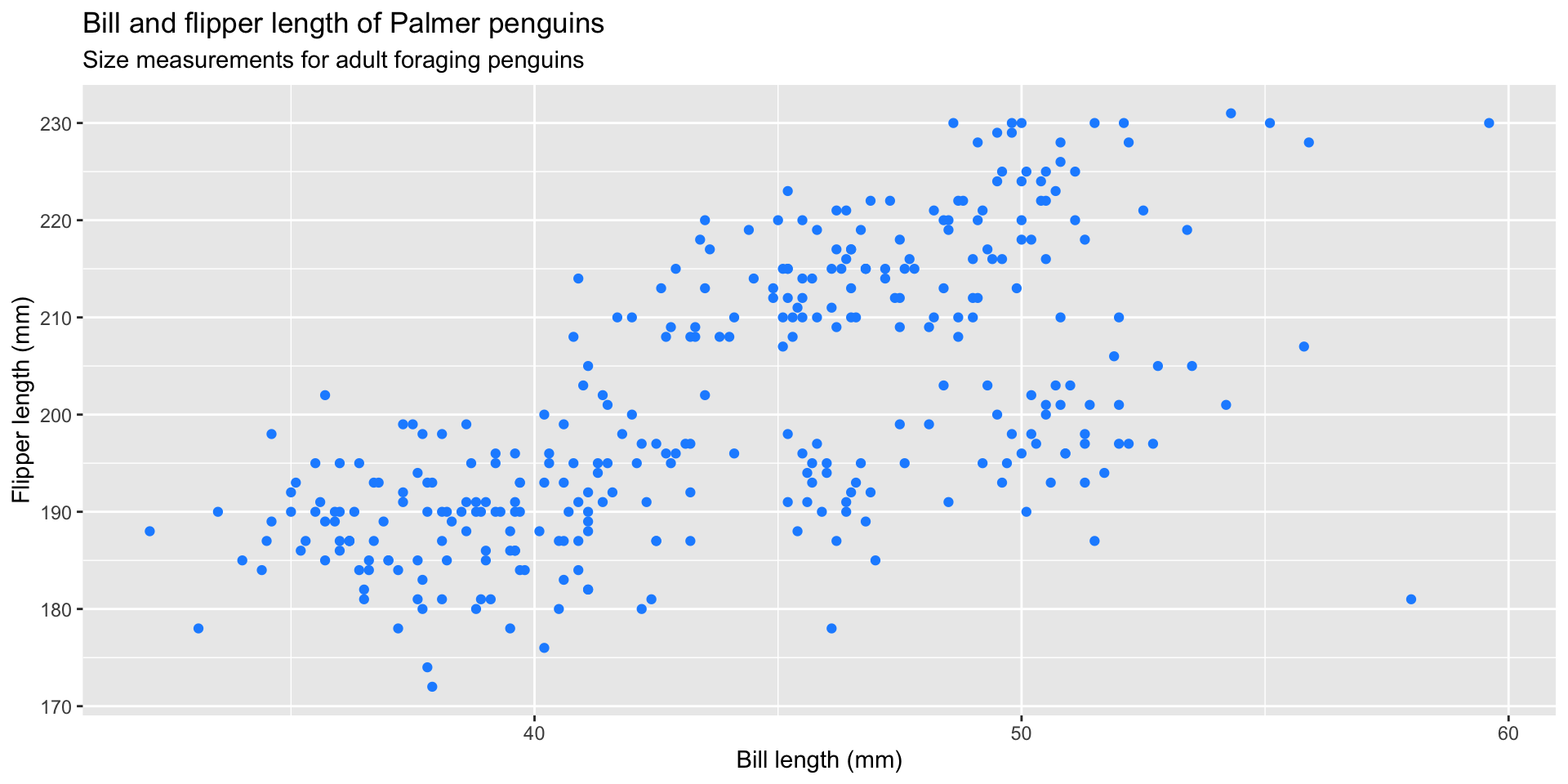
graph 12: Setting graph aesthetics
Change the code below to make the points "firebrick" red
Create labels
What color will the points be on this graph?

TIP: the legend tells us geom_point() is looking for a mapped variable in the penguins dataset named "firebrick"
Combining layers
graph 13: New layer, new data, no problem
Each geom_*() function also has a data argument, so we can supply new data at each layer
Create data label and source
big_penguins <- mutate(big_penguins,
label = case_when(
bill_length_mm == 59.6 ~ paste0("long bill = ", bill_length_mm),
bill_depth_mm == 21.5 ~ paste0("deep bill = ", bill_depth_mm),
flipper_length_mm == 231 ~ paste0("big flipper = ", flipper_length_mm),
body_mass_g == 6300 ~ paste0("big bird = ", body_mass_g)),
source = case_when(
bill_length_mm == 59.6 ~ "max bill length",
bill_depth_mm == 21.5 ~ "max bill depth",
flipper_length_mm == 231 ~ "max flipper length",
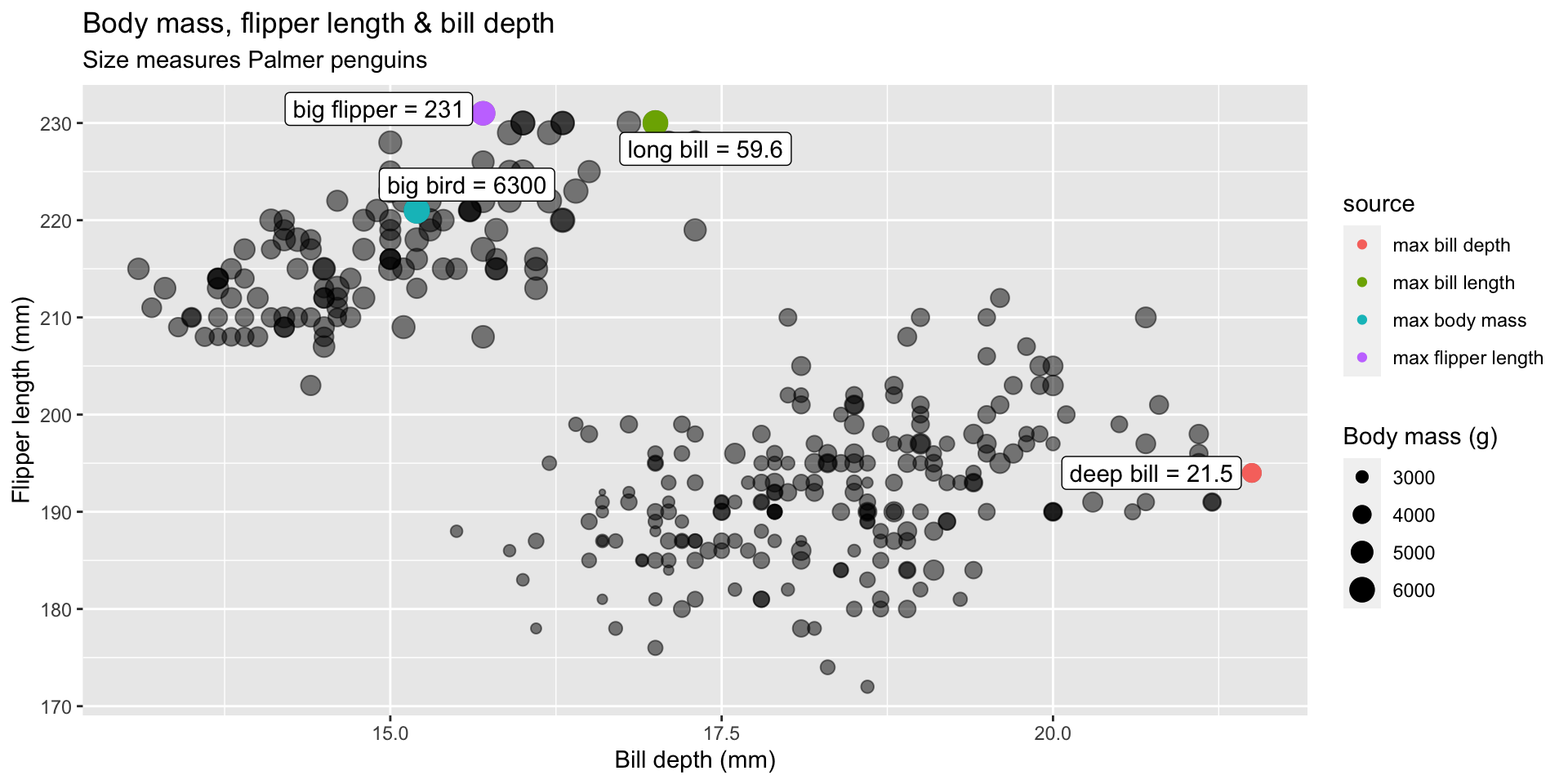
body_mass_g == 6300 ~ "max body mass"))Our label dataset
Objective: Create a scatter-plot to show the relationship between body mass, flipper length, and bill length.
| label | source |
|---|---|
| long bill = 59.6 | max bill length |
| deep bill = 21.5 | max bill depth |
| big flipper = 231 | max flipper length |
| big bird = 6300 | max body mass |
graph 13: Layer 1
Create layer 1 with penguins_no_miss data and geom_point()
Create labels
Assign x, y, size, and alpha

graph 14: Layer 2
Create layer 2 with another geom_point() using color and size
Use scale_size() to adjust point scaling

graph 15: Label 3 (max values)
Facets
Small multiples
From ggplot2 book
“Small multiples are a powerful tool for exploratory data analysis: you can rapidly compare patterns in different parts of the data and see whether they are the same or different.”
Facets = small multiples
In the previous graph, we used multiple aesthetics (color, size, shape)
Can we explore these relationships by sex or species?
Store graph 15 in ggp_penguin_measures

graph 16: Facet by sex
Use facet_wrap() to view our previous graph by sex
graph 17: Facet by species
Change facet_wrap() to build graphs by species and add theme
Change facet_wrap() to ~ species
Add theme_minimal() and labels

Recap
What we’ve covered
- Build labels (set your expectations)
- View data before building any graphs
- Building graphs layer-by-layer (data, mapping, geoms)
- Mapping variables to graph elements (color, position, size, etc.)
- Extending graphs by combining layers
- Using facets to explore relationships
Thanks!