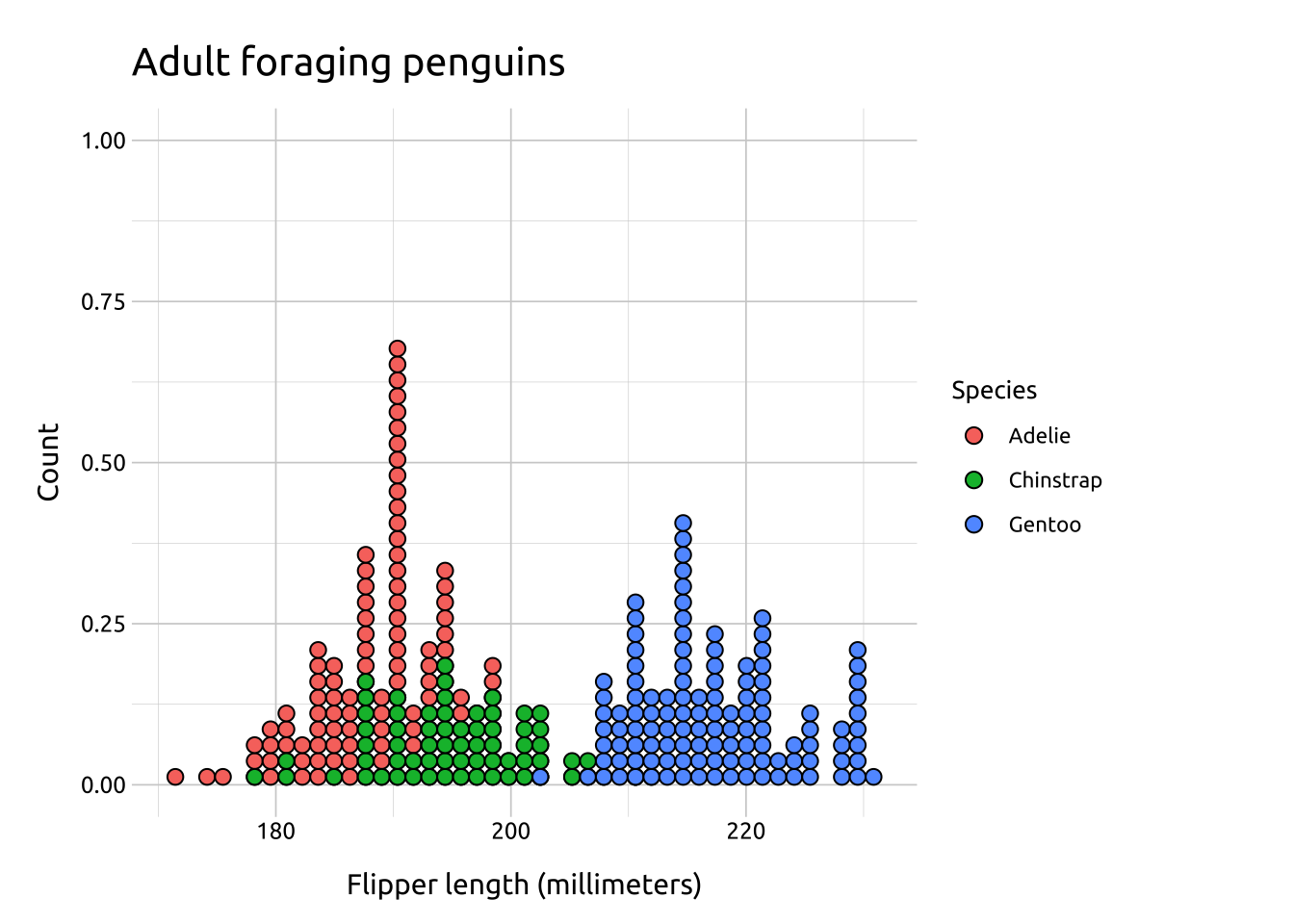
Overlapping dot plots
Description
Overlapping dot plots display distributions of a continuous variable across the levels of a categorical variable.
To adjust the dot plot display to look similar to a histogram or frequency polygon, change the method and binposition arguments.
Getting set up
PACKAGES:
Install packages.
Code
install.packages("palmerpenguins")
library(palmerpenguins)
library(ggplot2)DATA:

Remove missing sex from penguins
Code
peng_dotplot <- dplyr::filter(penguins, !is.na(sex))
dplyr::glimpse(peng_dotplot)Rows: 333
Columns: 8
$ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Adel…
$ island <fct> Torgersen, Torgersen, Torgersen, Torgersen, Torgerse…
$ bill_length_mm <dbl> 39.1, 39.5, 40.3, 36.7, 39.3, 38.9, 39.2, 41.1, 38.6…
$ bill_depth_mm <dbl> 18.7, 17.4, 18.0, 19.3, 20.6, 17.8, 19.6, 17.6, 21.2…
$ flipper_length_mm <int> 181, 186, 195, 193, 190, 181, 195, 182, 191, 198, 18…
$ body_mass_g <int> 3750, 3800, 3250, 3450, 3650, 3625, 4675, 3200, 3800…
$ sex <fct> male, female, female, female, male, female, male, fe…
$ year <int> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007…The grammar
binwidth= When method is “histodot”, this specifies bin width. Defaults to 1/30 of the range of the databinpositions= “all” determines positions of the bins with all the data taken together; this is used for aligning dot stacks across multiple groups.
CODE:
Create labels with labs()
Initialize the graph with ggplot() and provide data
Map flipper_length_mm to x
Map species to fill (inside factor())
Inside geom_dotplot, set method to "histodot", binwidth to 1.35, and binpositions to "all"
Code
labs_ovrlp_dotplot <- labs(
title = "Adult foraging penguins",
x = "Flipper length (millimeters)",
y = "Count",
fill = "Species")
ggp2_ovrlp_dotplot <- ggplot(data = peng_dotplot,
aes(x = flipper_length_mm,
fill = factor(species))) +
geom_dotplot(
method = "histodot",
binwidth = 1.35,
binpositions = "all")
ggp2_ovrlp_dotplot +
labs_ovrlp_dotplotGRAPH: