install.packages('ellmer')
# or the dev version
pak::pak('tidyverse/ellmer')29 gander
gander goes beyond ellmer and chores by extending the use of ellmer models to,
“talk[ing] to the objects in your R environment.”1
The previous LLM package we’ve covered required background information and conditions to be provided via prompts. gander is able to ‘peek around’ in our current R environment to provide framing and context.
29.1 Configuration
First we’ll install the ellmer and gander packages:
install.packages('gander')
# or the dev version
pak::pak("simonpcouch/gander")Configure the model in your .Renviron and .Rprofile files (an API key is required):
# configure ANTHROPIC_API_KEY, OPENAI_API_KEY, etc.
usethis::edit_r_environ() # configure .gander_chat
usethis::edit_r_profile()
options(
.gander_chat = ellmer::chat_anthropic()
)29.1.1 Keyboard shortcut
Configure the keyboard shortcut following the instructions on the package website (RStudio ![]() or Positron
or Positron ![]() ).
).
I use the recommended Ctrl+Cmd+G (or Ctrl+Alt+G on Windows).
29.3 Developing with gander
Launch app with the shinypak package:
launch('29_llm-gander')To develop with gander interactively, highlight the code you’d like to send to the model and use the keyboard shortcut:
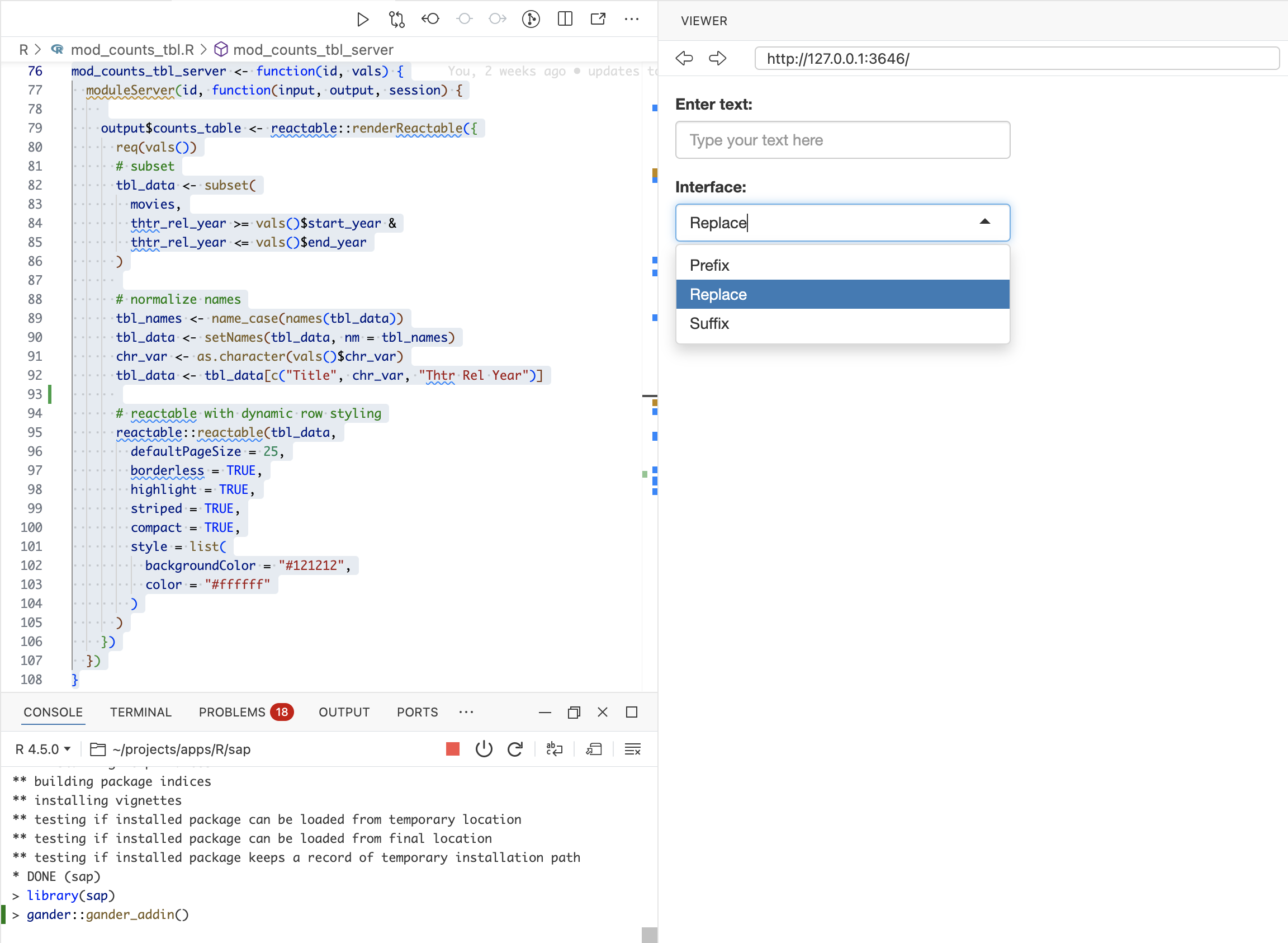
The gander addin includes a text box and a select input with Prefix, Replace, or Suffix.
29.3.1 Package functions
Lets assume we’ve made the decision to change the reactable tables to use the gt package. We’ll start by converting a single reactable table output (in R/mod_counts_tbl.R) to use a gt table.
After loading the new gt table function, we can test the code by launching our app:
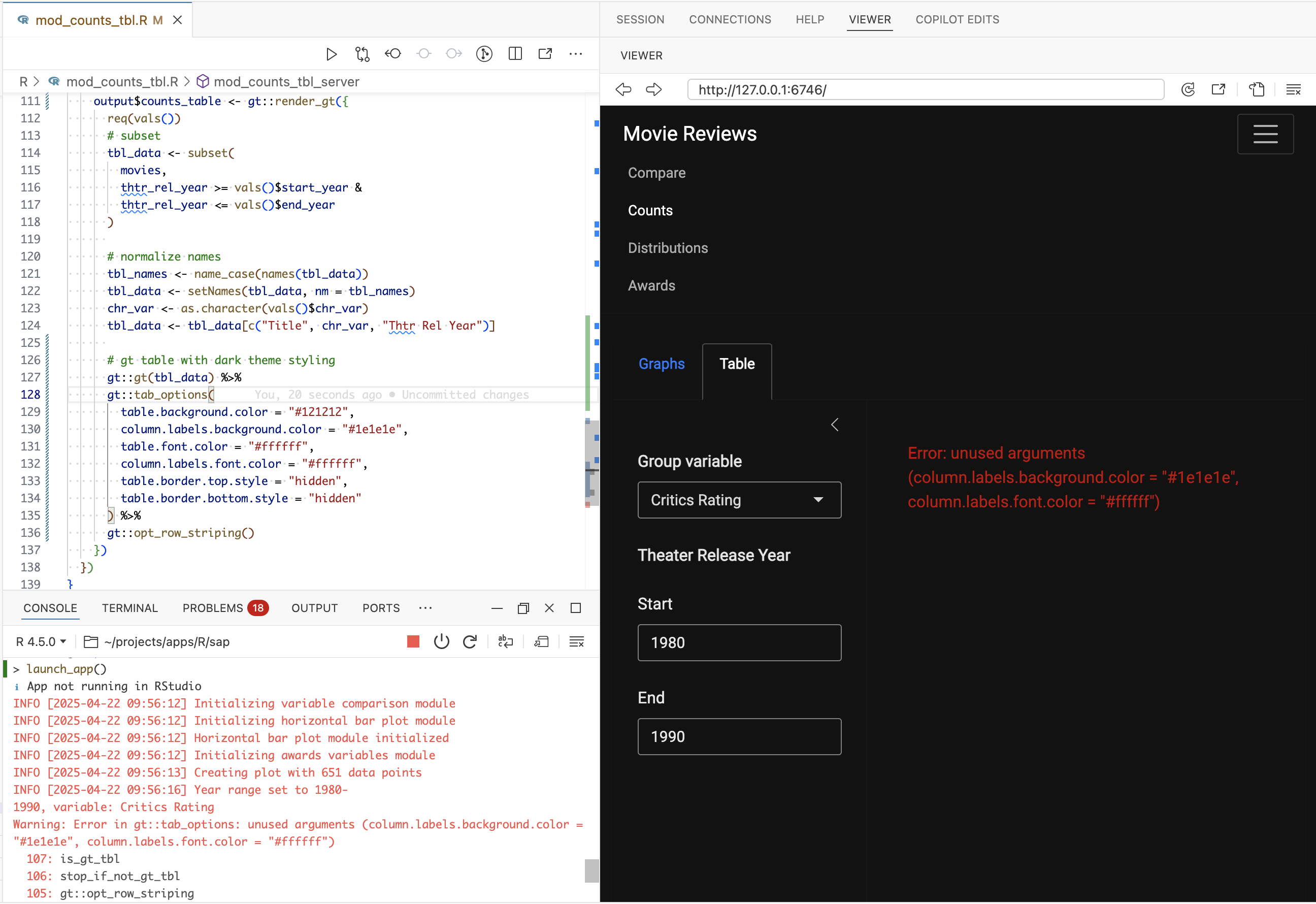
We can see the initial response throws an error, but we’ll use this as a opportunity to explore how the gander addin works.
29.4 Context
We can view the background/context passed to the model using gander::peek(). After giving us the model, turns, tokens and cost, the results are separated into system, user, and assistant.
<Chat Anthropic/claude-3-7-sonnet-latest turns=3 tokens=1178/336 $0.01>29.4.1 System
The system portion tells us how each response is framed and provides, ‘additional instructions to the model, shaping its responses to your needs.’4
── system [0] ────────────────────────────────────────────────────────────
You are a helpful but terse R data scientist. Respond only with valid R code:
no exposition, no backticks. Always provide a minimal solution and refrain
from unnecessary additions. Use tidyverse style and, when relevant, tidyverse
packages. For example, when asked to plot something, use ggplot2, or when
asked to transform data, using dplyr and/or tidyr unless explicitly
instructed otherwise. I like to think of system prompts as an assistant’s ‘personality,’ because it affects every response.
29.4.2 User
The user prompt section gives the model information on the R environment, file contents, etc. In this example we can see it provided the entire module (UI and server functions) and roxygen2 documentation.
Note the ‘Up to this point, the contents of my r file reads: ’ and ’Now, convert the reactable output to a gt table: ” delineation.
── user [1178] ─────────────────────────────────────────────────────────────
Up to this point, the contents of my r file reads:
\`\`\`r
#' UI for counts table module
#'
#' Creates a reactive table displaying count data. This function is designed
#' to work together with [mod_counts_tbl_server()].
#'
#' @param id A character string used to identify the namespace for the module.
#'
#' @return A `tagList` containing UI elements:
#' * A reactive table output that displays the count data
#'
#' @seealso [mod_counts_tbl_server()] for the server-side logic
#'
#' @examples
#' # UI implementation
#' ui <- fluidPage(
#' mod_counts_tbl_ui("counts1")
#' )
#'
#' # Server implementation
#' server <- function(input, output, session) {
#' mod_counts_tbl_server("counts1", counts_data = reactive(data))
#' }
#'
mod_counts_tbl_ui <- function(id) {
ns <- NS(id)
tagList(
reactable::reactableOutput(
outputId = ns("counts_table")
)
)
}
#' Server function for the count table module
#'
#' Creates a reactive table showing movies based on selected filters. This
#' function is designed to work together with a corresponding UI function.
#'
#' @param id A character string used to identify the namespace for the module.
#' @param vals A reactive expression that returns a list containing at least:
#' * `start_year`: numeric value for the earliest year to include
#' * `end_year`: numeric value for the latest year to include
#' * `chr_var`: symbol representing the variable to display alongside title
#'
#' @return Creates the following reactive elements within the module's namespace:
#' * `counts_table`: A reactive Reactable table with three columns:
#' - Title: The movie title
#' - The selected character variable from `vals()$chr_var`
#' - Thtr Rel Year: The theatrical release year
#'
#' The table includes styling with a dark background, white text, and features
#' such as highlighting, striping, and compact display.
#'
#' @details
#' The function filters the global `movies` dataset based on the year range
#' provided in `vals()`. Column names are normalized using the `name_case()`
#' function before displaying.
#'
#' @seealso [mod_counts_tbl_()] The corresponding UI function for this module
#'
#' @examples
#' # Server implementation
#' server <- function(input, output, session) {
#' # Create a reactive values list
#' selected_vals <- reactive({
#' list(
#' start_year = 2000,
#' end_year = 2010,
#' chr_var = sym("Critics Rating")
#' )
#' })
#'
#' # Call the module server
#' mod_counts_tbl_server("movie_table", selected_vals)
#' }
#'
\`\`\`
Now, convert the reactable output to a gt table:
\`\`\`r
mod_counts_tbl_server <- function(id, vals) {
moduleServer(id, function(input, output, session) {
output$counts_table <- reactable::renderReactable({
req(vals())
# subset
count_data <- subset(
movies,
thtr_rel_year >= vals()$start_year &
thtr_rel_year <= vals()$end_year
)
# Convert selected variable to snake_case
group_var <- name_case(as.character(vals()$chr_var), case = "lower")
# Count per group using tidy evaluation
tbl_data <- count_data |>
dplyr::group_by(.data[[group_var]]) |>
dplyr::summarise(n = dplyr::n(), .groups = "drop") |>
dplyr::arrange(dplyr::desc(n))
# Normalize column names in the data
names(tbl_data) <- name_case(names(tbl_data))
# reactable with dynamic row styling
reactable::reactable(tbl_data,
defaultPageSize = 25,
borderless = TRUE,
highlight = TRUE,
striped = TRUE,
compact = TRUE,
style = list(
backgroundColor = "#121212",
color = "#ffffff"
)
)
})
})
}
\`\`\`29.4.3 Assistant
Finally, the assistant is the response from the model (given the system and user prompts).
── assistant [336] ───────────────────────────────────────────────────────
mod_counts_tbl_server <- function(id, vals) {
moduleServer(id, function(input, output, session) {
output$counts_table <- gt::render_gt({
req(vals())
# subset
count_data <- subset(
movies,
thtr_rel_year >= vals()$start_year &
thtr_rel_year <= vals()$end_year
)
# Convert selected variable to snake_case
group_var <- name_case(as.character(vals()$chr_var), case = "lower")
# Count per group using tidy evaluation
tbl_data <- count_data |>
dplyr::group_by(.data[[group_var]]) |>
dplyr::summarise(n = dplyr::n(), .groups = "drop") |>
dplyr::arrange(dplyr::desc(n))
# Normalize column names in the data
names(tbl_data) <- name_case(names(tbl_data))
# gt table with dark theme styling
gt::gt(tbl_data) %>%
gt::tab_options(
table.background.color = "#121212",
column.labels.background.color = "#1e1e1e",
table.font.color = "#ffffff",
column.labels.font.color = "#ffffff",
table.border.top.style = "hidden",
table.border.bottom.style = "hidden"
) %>%
gt::opt_row_striping()
})
})
}29.4.4 In practice
The context gander provides is enough information to 1) ensure the response is aware of the corresponding UI function, and 2) notice we’re developing an R package (i.e., use explicit namespacing with pkg::fun()).
The errors we’re encountering are due to incorrect arguments passed to gt::tab_options():
gt::tab_options(
table.background.color = "#121212",
column.labels.background.color = "#1e1e1e",
table.font.color = "#ffffff",
column.labels.font.color = "#ffffff",
table.border.top.style = "hidden",
table.border.bottom.style = "hidden"
) - 1
-
Should be
column_labels.background.color
- 2
-
Doesn’t exist (uses
table.font.color)
After making the changes to the server-side gt table code, we need to remember to update the corresponding UI code with the gt rendering function (recall that we didn’t specify any changes to this function in the prompt).
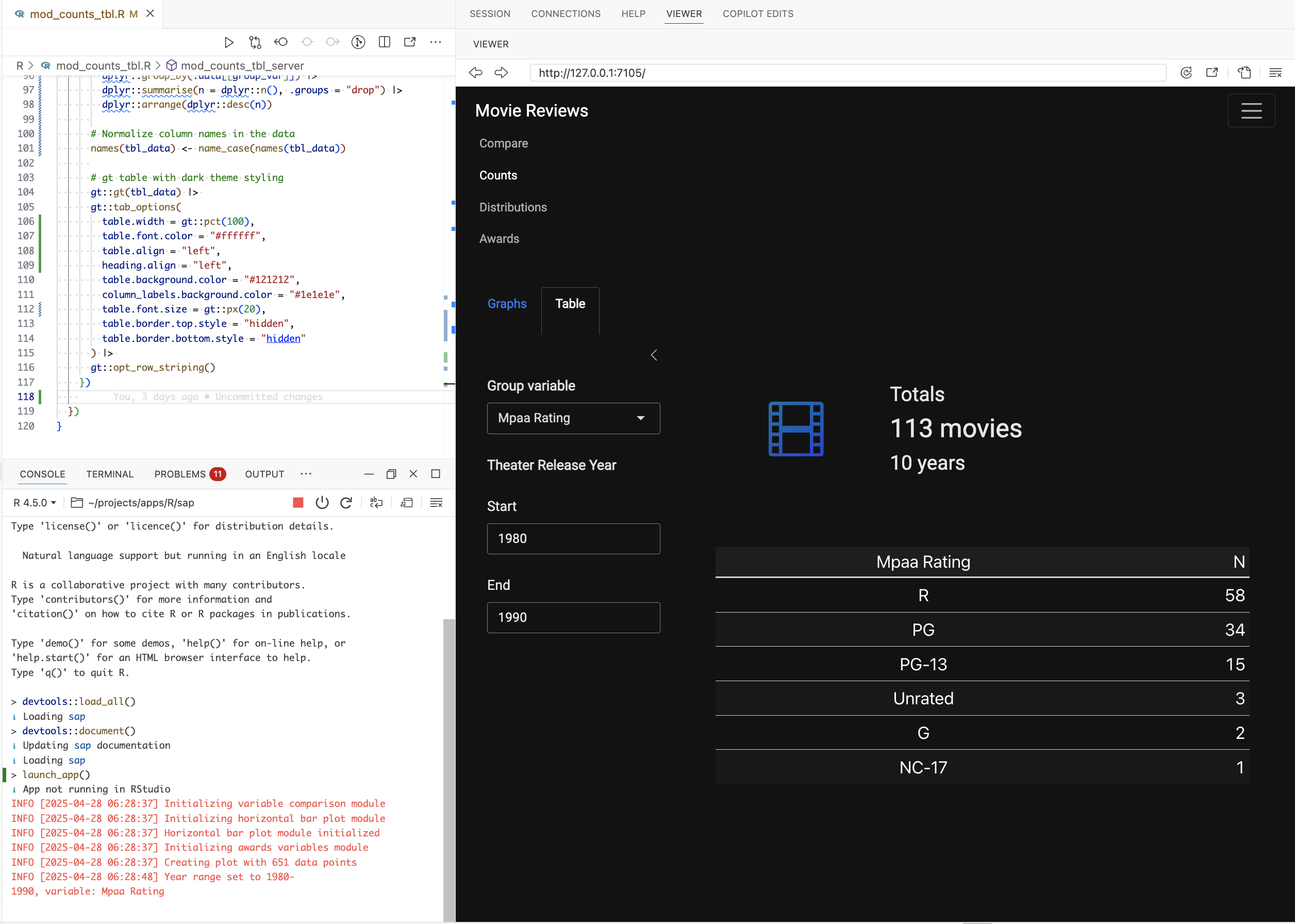
After updating the UI module function and loading the changes, we can see the Counts table below:
29.5 Graph adjustments
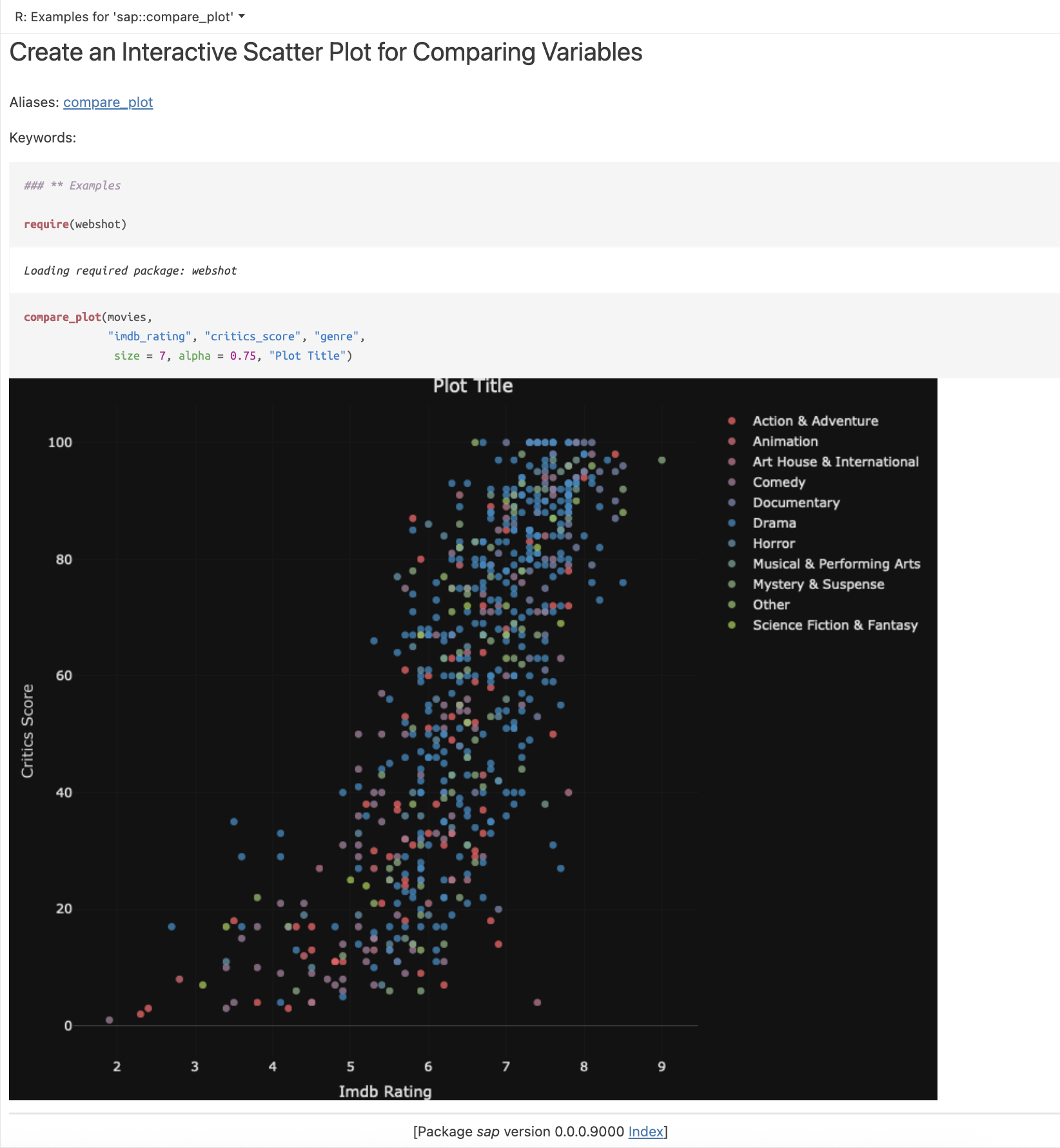
I’ve found the gander addin to be incredibly helpful for visualization development in Shiny modules. In the Compare tab, we display a plotly scatter plot. I’ve asked gander to convert the plotly code in the module server function into a utility function (R/compare_plot.R):
Prompt: “Convert the plotly code inside renderPlotly() into a separate utility function (compare_plot()).”
This resulted in the utility function below:
show/hide R/compare_plot.R
#' Create an Interactive Scatter Plot for Comparing Variables
#'
#' This function creates an interactive scatter plot using plotly to compare two
#' variables with points colored by a third variable.
#'
#' @param data A data frame containing the variables to plot
#' @param x Character string with the name of the variable to plot on the x-axis
#' @param y Character string with the name of the variable to plot on the y-axis
#' @param color Character string with the name of the variable to use for
#' coloring points
#' @param size Numeric value for the size of the points (default: 5)
#' @param alpha Numeric value for the transparency of the points (default: 0.7)
#' @param title Character string for the plot title. If NULL, a default title is
#' generated
#'
#' @return a plotly interactive scatter plot object
#'
#' @examples
#' require(webshot)
#' compare_plot(movies,
#' "imdb_rating", "critics_score", "genre",
#' size = 7, alpha = 0.75, "Plot Title")
#'
#' @export
#'
compare_plot <- function(data, x, y, color, size = 5, alpha = 0.7, title = NULL) {
# title if not provided
if (is.null(title)) {
title <- paste(name_case(x), "vs.", name_case(y))
}
# plot
plot <- plotly::plot_ly(
data = data,
x = ~get(x),
y = ~get(y),
color = ~get(color),
text = ~title,
type = "scatter",
mode = "markers",
colors = clr_pal3,
marker = list(
size = size,
opacity = alpha
)
) |>
# title
plotly::layout(
title = list(
text = title,
font = list(color = "#e0e0e0")
),
# x-label
xaxis = list(
title = name_case(x),
titlefont = list(color = "#e0e0e0"),
tickfont = list(color = "#e0e0e0")
),
# y-label
yaxis = list(
title = name_case(y),
titlefont = list(color = "#e0e0e0"),
tickfont = list(color = "#e0e0e0")
),
# legend
legend = list(
font = list(color = "#e0e0e0")
),
# background color
plot_bgcolor = "#121212",
paper_bgcolor = "#121212"
)
return(plot)
}Placing the graph code in a separate utility function can make it easier to debug/write tests. Writing graph examples in the roxygen2 documentation also makes it possible to view minor changes to the graph without dealing with the reactivity inside the application:
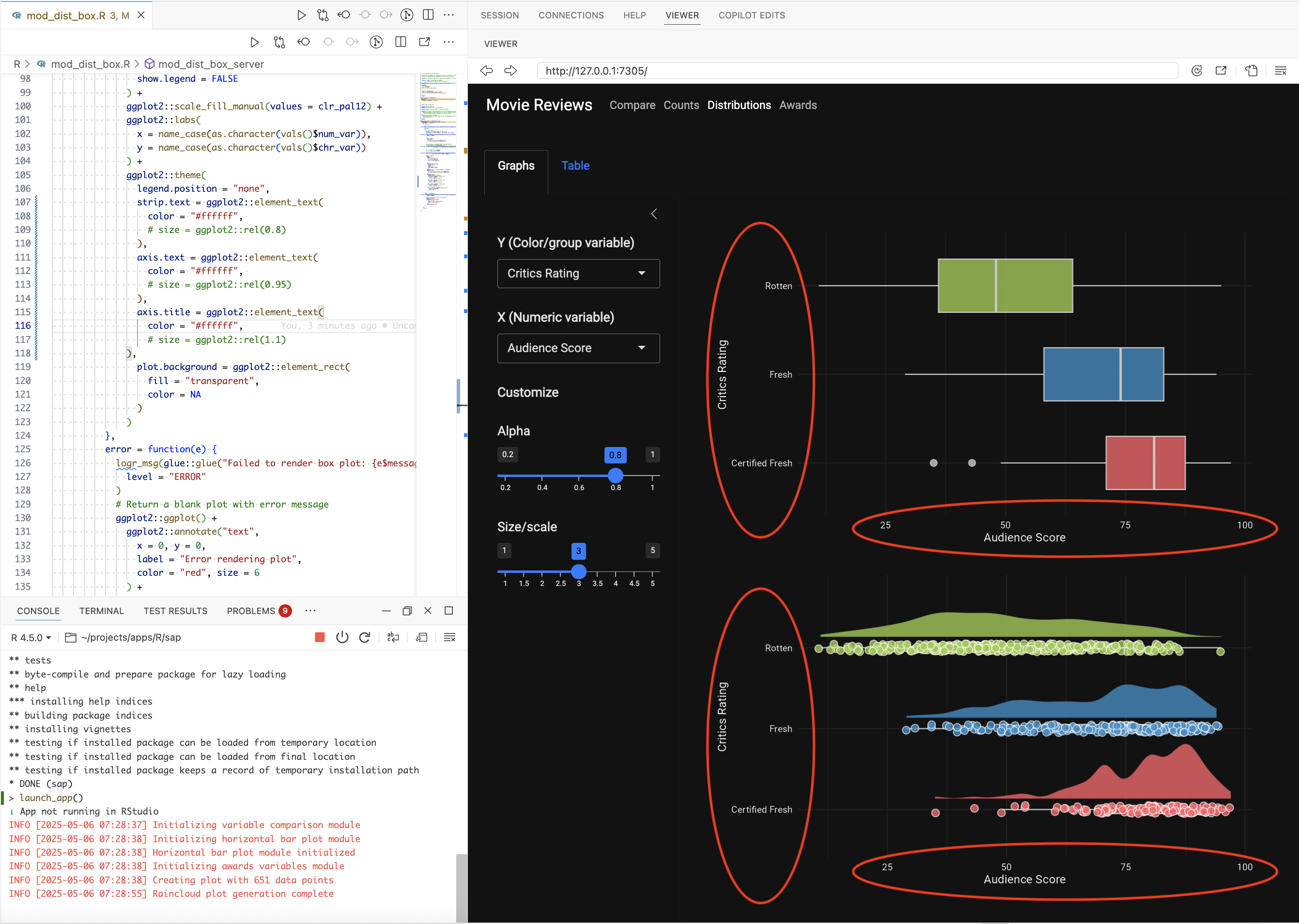
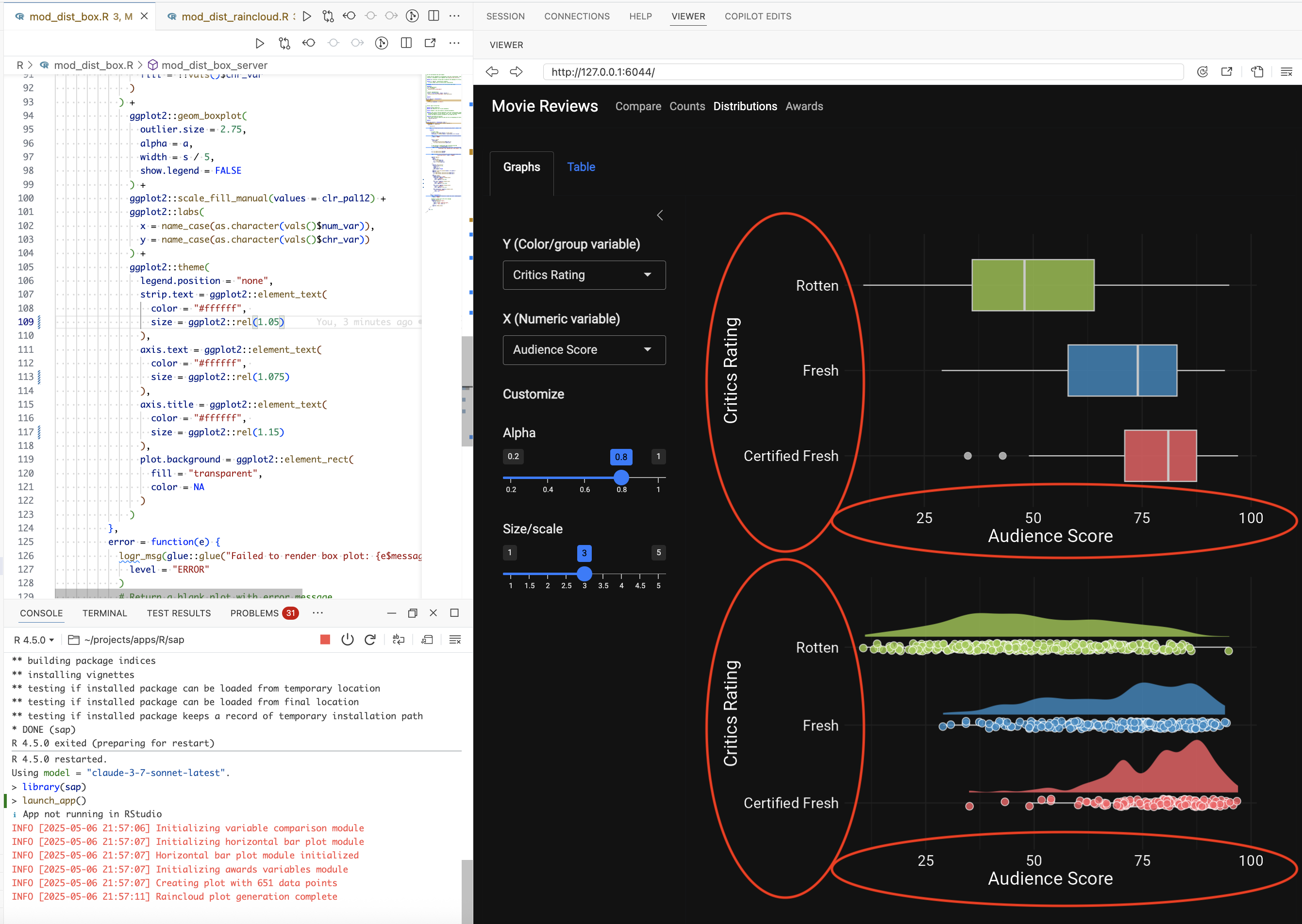
I also noticed the text size was too small in the graphs on the Distribution tabs, so I also asked gander to make the font size in the graph adjust with the display:
Prompt: “Make the font in the ggplot2 graphs adjust to the screen size”
gander only returned a few changes to the ggplot2 code, but these small changes represent a lot saved time because there are so many ggplot2::theme() arguments (and they usually take multiple iterations to get correct):
ggplot2::theme(
strip.text = ggplot2::element_text(
color = "#ffffff",
size = ggplot2::rel(1.05)
),
axis.text = ggplot2::element_text(
color = "#ffffff",
size = ggplot2::rel(1.075)
),
axis.title = ggplot2::element_text(
color = "#ffffff",
size = ggplot2::rel(1.15)
)
)Graph changes like these can take up a lot of time if we’re reading through package documentation (or Stack Overflow/Posit Community). But gander can handle tasks like this quickly, although a foundational understanding of plotly and ggplot2 makes it easier to debug any errors the generated code produces.
29.6 Downloadable reports
I wanted to stretch the functionality of gander, so I asked the chatbot to create a downloadable report module. This required a series of prompts to get working, but I think it serves as a great example of using the gander addin to iterate through a solution/feature.
First, I created the report template file (inst/rmd/report_template.Rmd) with the following YAML:
---
title: "Movie Analysis Report"
date: "December 08, 2025"
output:
html_document:
toc: true
toc_depth: 2
theme: united
params:
compare_values: NA
count_values: NA
dist_values: NA
award_values: NA
---The params in the report will be passed from the new R/mod_report.R module, which we can ask gander to generate by highlighting the contents of app’s server function (nav_server()) and entering the following prompt:
Prompt: Create a module for downloading an R Markdown HTML report using the
_valuesas parameters.
The resulting module is below:
show/hide mod_report module
#' Report download UI
#'
#' @param id Module ID
#'
#' @return UI elements for downloading report
#'
#' @export
mod_report_ui <- function(id) {
ns <- NS(id)
tagList(
downloadButton(ns("download_report"), "Download Report")
)
}
#' Generate and download R Markdown report
#'
#' @param id Module ID
#' @param scatter_values Scatter plot reactive values
#' @param count_values Count data reactive values
#' @param dist_values Distribution reactive values
#' @param award_values Awards reactive values
#'
#' @return None
#'
#' @export
mod_report_server <- function(id, scatter_values, count_values, dist_values, award_values) {
moduleServer(id, function(input, output, session) {
output$download_report <- downloadHandler(
filename = function() {
paste("report-", Sys.Date(), ".html", sep = "")
},
content = function(file) {
# create a temporary directory for report generation
tempDir <- tempdir()
tempReport <- file.path(tempDir, "report.Rmd")
# copy the report template to the temp directory
file.copy(system.file("rmd", "report_template.Rmd", package = "sap"),
tempReport, overwrite = TRUE)
# set up parameters to pass to Rmd
params <- list(
compare_values = scatter_values(),
count_values = count_values(),
dist_values = dist_values(),
award_values = award_values()
)
# render the report
rmarkdown::render(
input = tempReport,
output_file = file,
params = params,
envir = new.env(parent = globalenv()))
}
)
})
}As we can see, the new module conforms to our naming convention and follows the guidance from the official generating reports documentation (i.e., it has a downloadHandler() with filename and content, and passes the params to rmarkdown::render()).
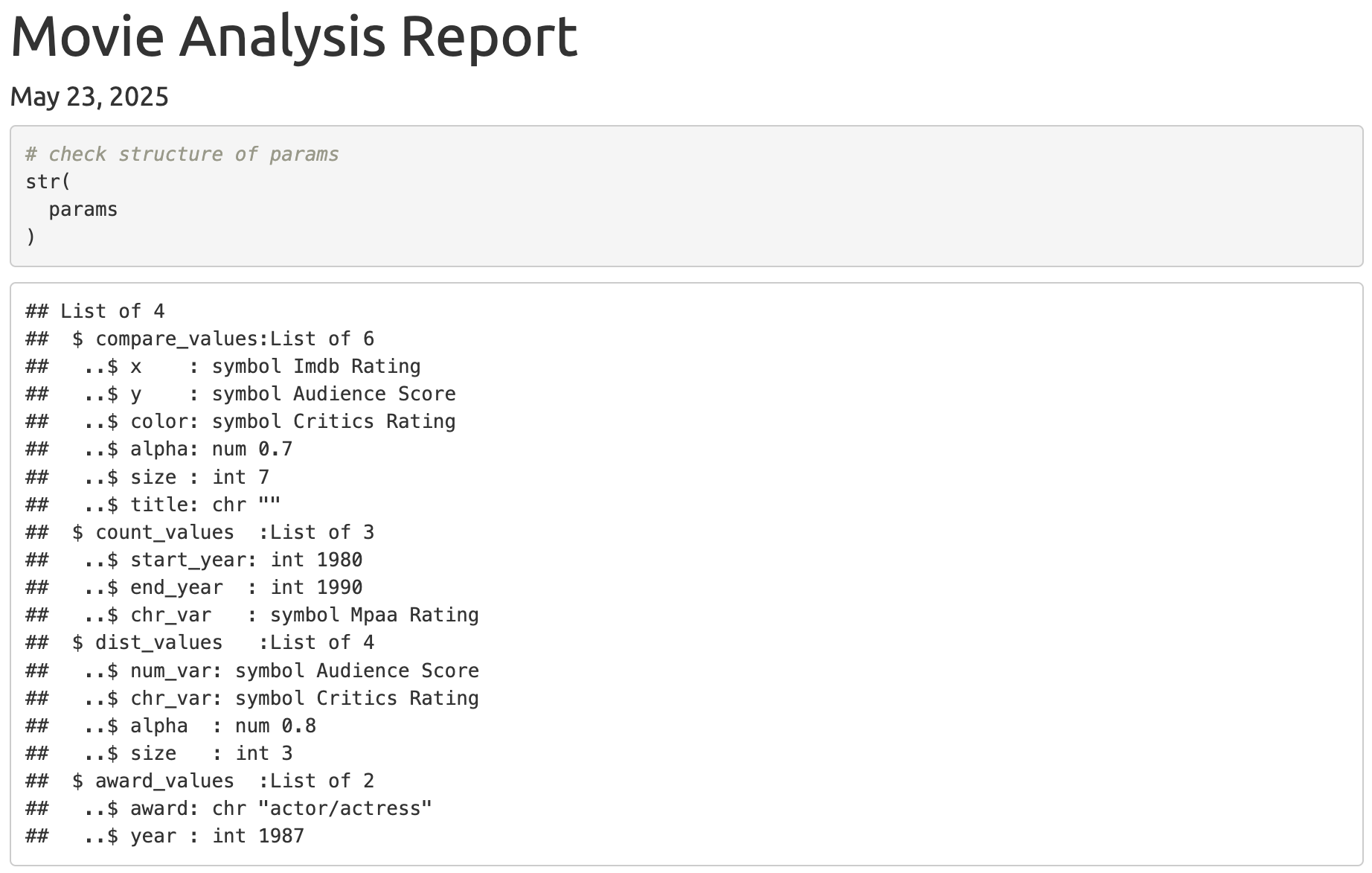
After implementing this module in the UI and the server, I can see the params have been passed to the .Rmd file by printing it’s structure:
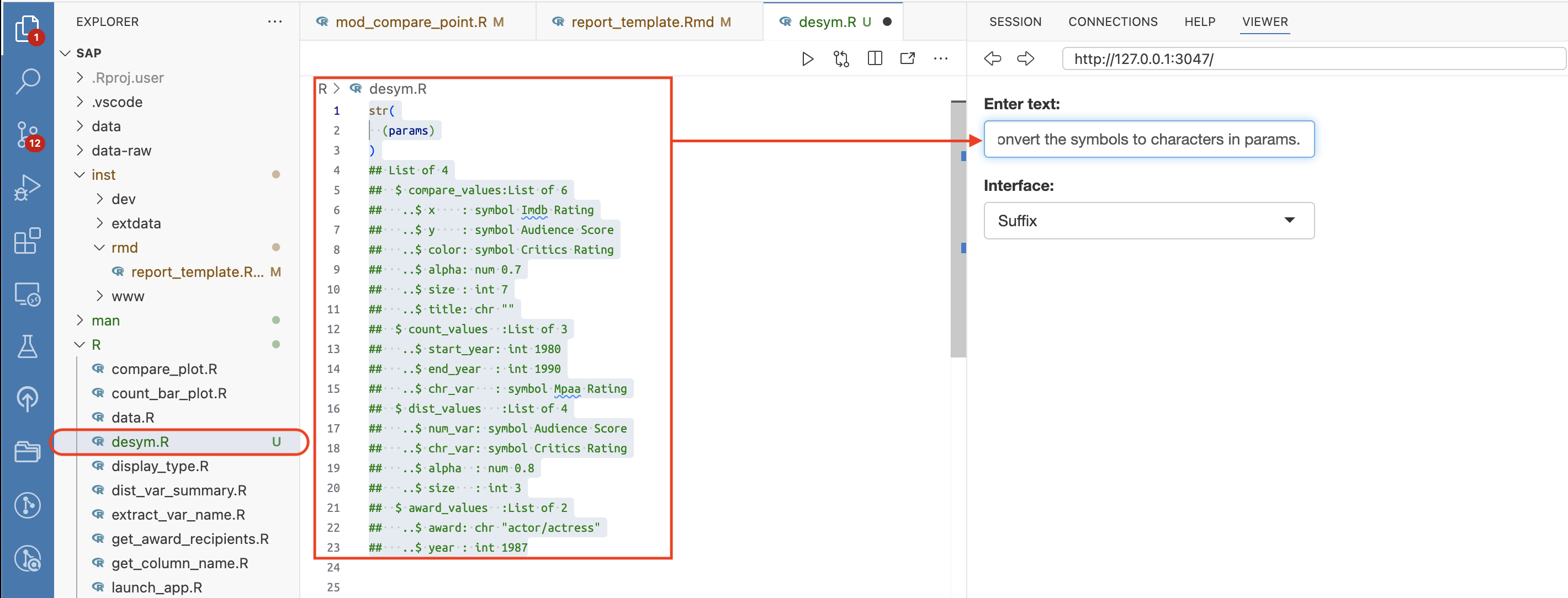
To convert the symbols to characters in params, I created an empty .R script (R/desym.R), pasted and highlighted the params structure output from the report, and asked gander to write a utility function:
Prompt: Write a
desymfunction that will convert the symbols to characters inparams.
show/hide desym() utility function
#' Convert symbols to character
#'
#' @param params_list
#'
#' @returns list of params with character values
#'
#' @export
#'
desym <- function(params_list) {
convert_symbol <- function(x) {
if (is.symbol(x)) {
return(as.character(x))
} else if (is.list(x)) {
return(lapply(x, convert_symbol))
} else {
return(x)
}
}
lapply(params_list, convert_symbol)
}We’ll add the desym() function to the mod_report_server() function to ensure only character values are passed to the report.
show/hide mod_report_server() with dsym()
# Set up parameters to pass to Rmd
params <- list(
compare_values = scatter_values(),
count_values = count_values(),
dist_values = dist_values(),
award_values = award_values()
)
# remove the symbols
clean_params <- desym(params)
# Render the report
rmarkdown::render(
input = tempReport,
output_file = file,
params = clean_params,
envir = new.env(parent = globalenv()))Now we have parameter values we can use with our new custom plotting function (compare_plot()). We can include the following in a R code block in inst/report_template.Rmd:
show/hide compare_plot() in inst/report_template.Rmd
tryCatch({
# extract variable names
x_var <- name_case(params$compare_values$x, case = "lower")
y_var <- name_case(params$compare_values$y, case = "lower")
color_var <- name_case(params$compare_values$color, case = "lower")
# create data
compare_data <- sap::movies
# create plot
compare_plot(data = compare_data,
x = x_var,
y = y_var,
color = color_var,
size = params$compare_values$size,
alpha = params$compare_values$alpha,
title = params$compare_values$title)
},
error = function(e) {
cat("\n\n**Error creating plot:**\n\n", e$message)
}
)- 1
-
Convert these to
lower_snake_caseto match the names inmovies
- 2
-
Can pass the original
paramsvalues unchanged.
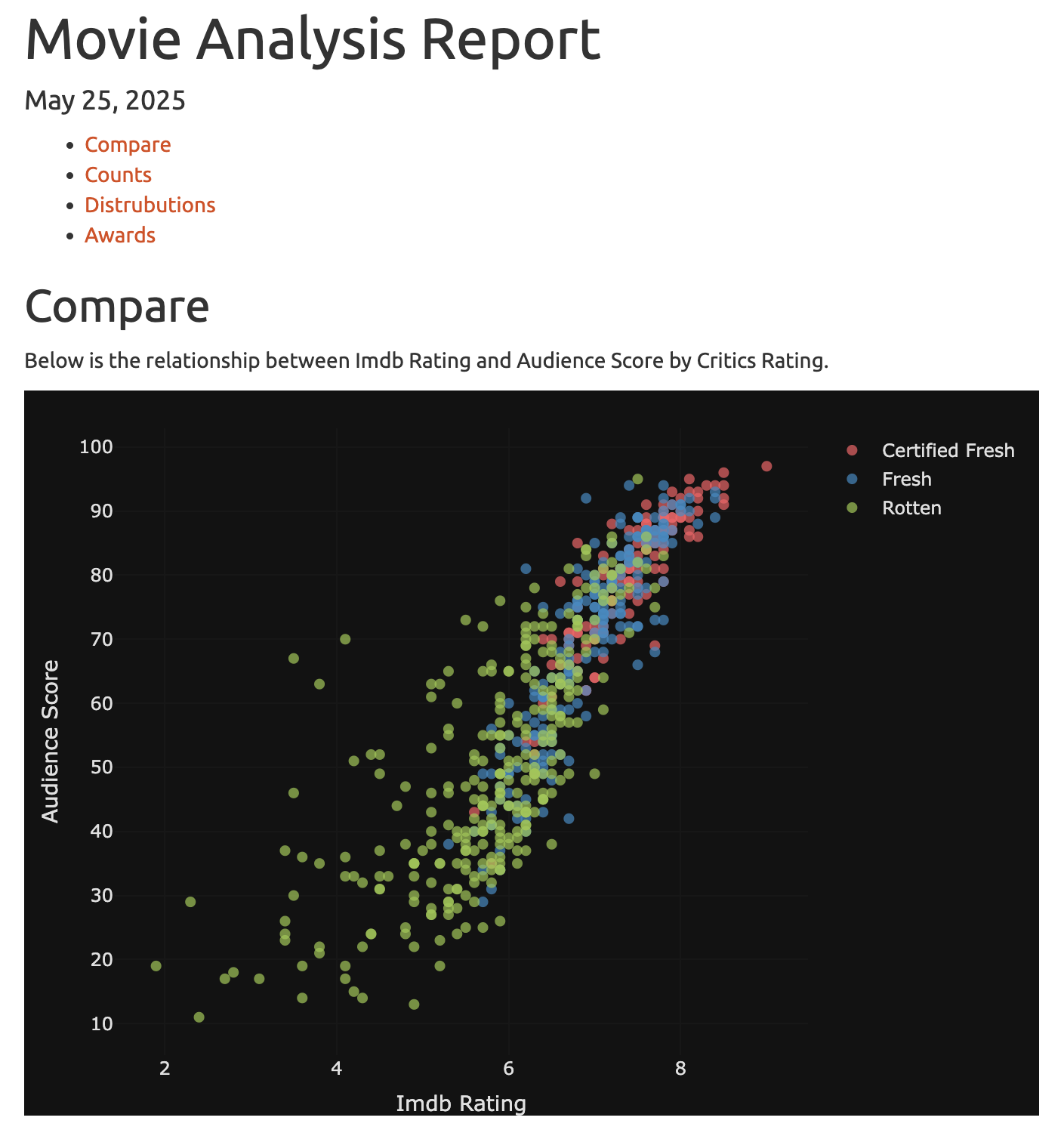
Our downloaded HTML report contains the following:
We’ll stop development here, but as you can see, the gander addin is incredibly helpful for editing modules, developing utility functions, and adding content to our downloadable R Markdown report.
Recap
ganderis the third LLM R package we’ve covered by developer Simon Couch (we also coveredensurein Section 16.1 andchoresin Chapter 28). If you’re interested in staying up on LLMs and their use in R package development, I highly recommend regularly checking his blog.↩︎This application uses the
page_navbarlayout frombslib. Review it’s structure in Section 28.1.↩︎See the Section 28.4.1 and previous Section 13.3 sections.↩︎
Read more about this in the What is a prompt? section of the
ellmerdocumentation.↩︎