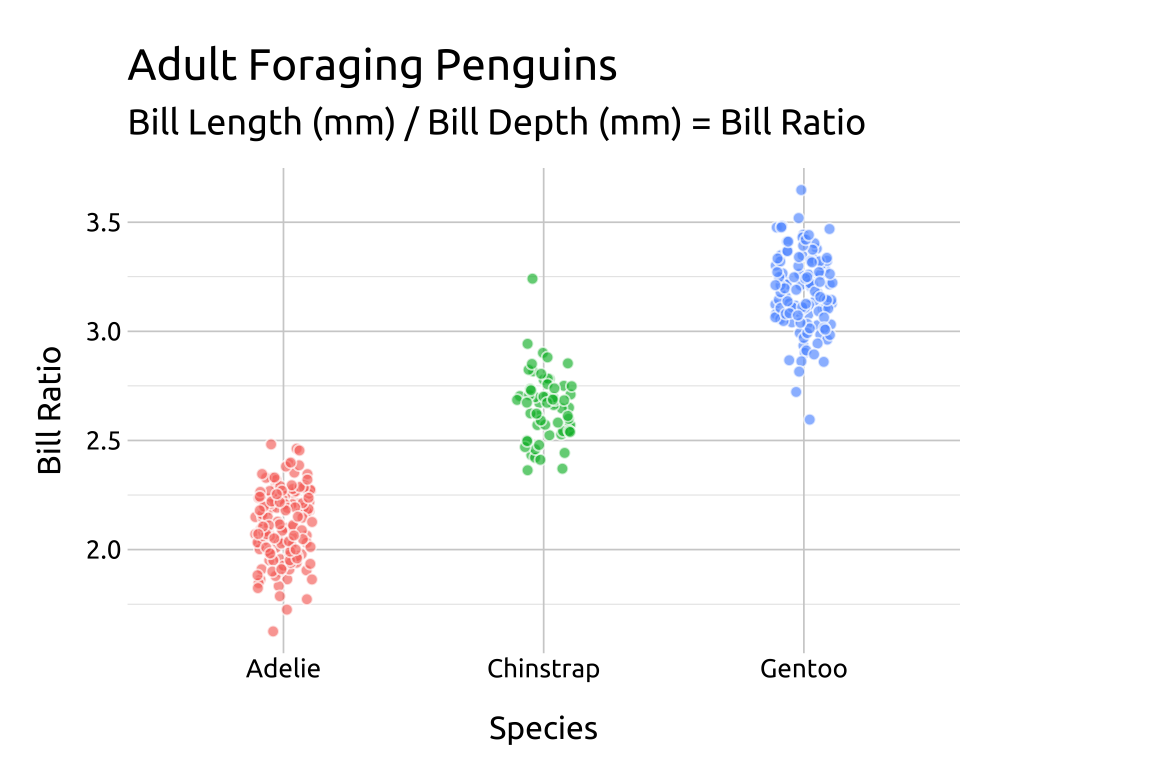
22 Beeswarm plots
22.1 Description
The beeswarm plot uses points to display the distribution of a continuous variable across the levels of a categorical variable.
The points are grouped by level, and the shape (or swarm) of the distribution is mirrored above and below the quantitative axis (similar to a violin plot).
We can create beeswarm plot using geom_jitter() or the ggbeeswarm package.
22.2 Set up
PACKAGES:
Install packages.
show/hide
# pak::pak("eclarke/ggbeeswarm")
library(ggbeeswarm)
install.packages("palmerpenguins")
library(palmerpenguins)
library(ggplot2)DATA:

Create peng_beeswarm by grouping penguins by species, then calculating the bill_ratio (bill_length_mm / bill_depth_mm), and then removing any missing values from bill_ratio
show/hide
peng_beeswarm <- palmerpenguins::penguins |>
dplyr::group_by(species) |>
dplyr::mutate(bill_ratio = bill_length_mm / bill_depth_mm) |>
dplyr::filter(!is.na(bill_ratio)) |>
dplyr::ungroup()
glimpse(peng_beeswarm)
#> Rows: 342
#> Columns: 9
#> $ species <fct> Adelie, Adelie, Adelie…
#> $ island <fct> Torgersen, Torgersen, …
#> $ bill_length_mm <dbl> 39.1, 39.5, 40.3, 36.7…
#> $ bill_depth_mm <dbl> 18.7, 17.4, 18.0, 19.3…
#> $ flipper_length_mm <int> 181, 186, 195, 193, 19…
#> $ body_mass_g <int> 3750, 3800, 3250, 3450…
#> $ sex <fct> male, female, female, …
#> $ year <int> 2007, 2007, 2007, 2007…
#> $ bill_ratio <dbl> 2.090909, 2.270115, 2.…22.3 Grammar
CODE:
Create labels with
labs()Initialize the graph with
ggplot()and providedataMap
speciesto thexaxis andcolorMap
bill_ratioto theyaxisAdd the
ggbeeswarm::geom_beeswarm()layer (withalpha)Remove the legend with
show.legend = FALSE
show/hide
labs_beeswarm <- labs(
title = "Adult Foraging Penguins",
subtitle = "Bill Length (mm) / Bill Depth (mm) = Bill Ratio",
x = "Species",
y = "Bill Ratio")
ggp2_beeswarm <- ggplot(data = peng_beeswarm,
aes(x = species,
y = bill_ratio,
color = species)) +
ggbeeswarm::geom_beeswarm(alpha = 2 / 3,
show.legend = FALSE)
ggp2_beeswarm +
labs_beeswarmGRAPH:
Adjust the size/shape of the swarm using method = or the geom_quasirandom() function
22.4 More info
Below is some additional arguments and methods for beeswarm plots.
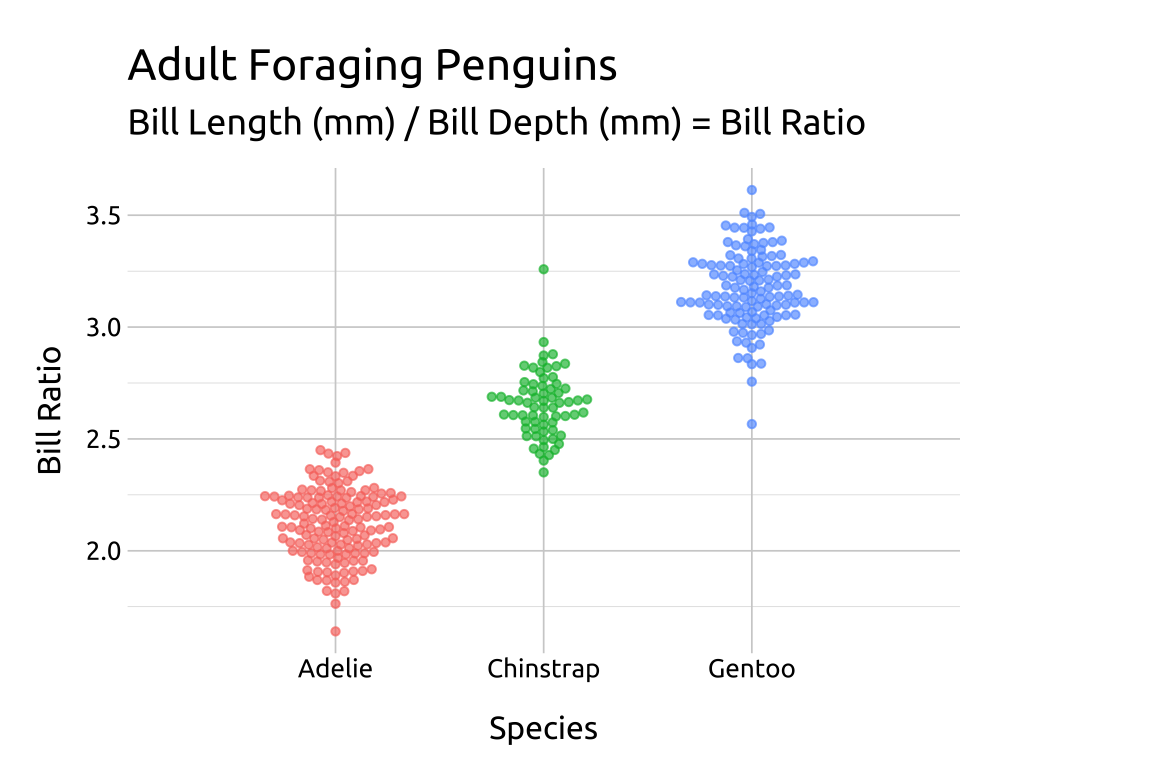
22.4.1 method
Use method to adjust the shape of the beeswarm (swarm, compactswarm, hex, square, center, or centre)
Set the point shape to 21 to control the fill and color
show/hide
ggp2_compact_swarm <- ggplot(data = peng_beeswarm,
mapping = aes(x = species,
y = bill_ratio,
color = species)) +
ggbeeswarm::geom_beeswarm(
aes(fill = species),
method = 'compactswarm',
dodge.width = 0.5,
shape = 21,
color = "#ffffff",
alpha = 2/3, size = 1.7,
show.legend = FALSE)
ggp2_compact_swarm +
# add labels
labs_beeswarm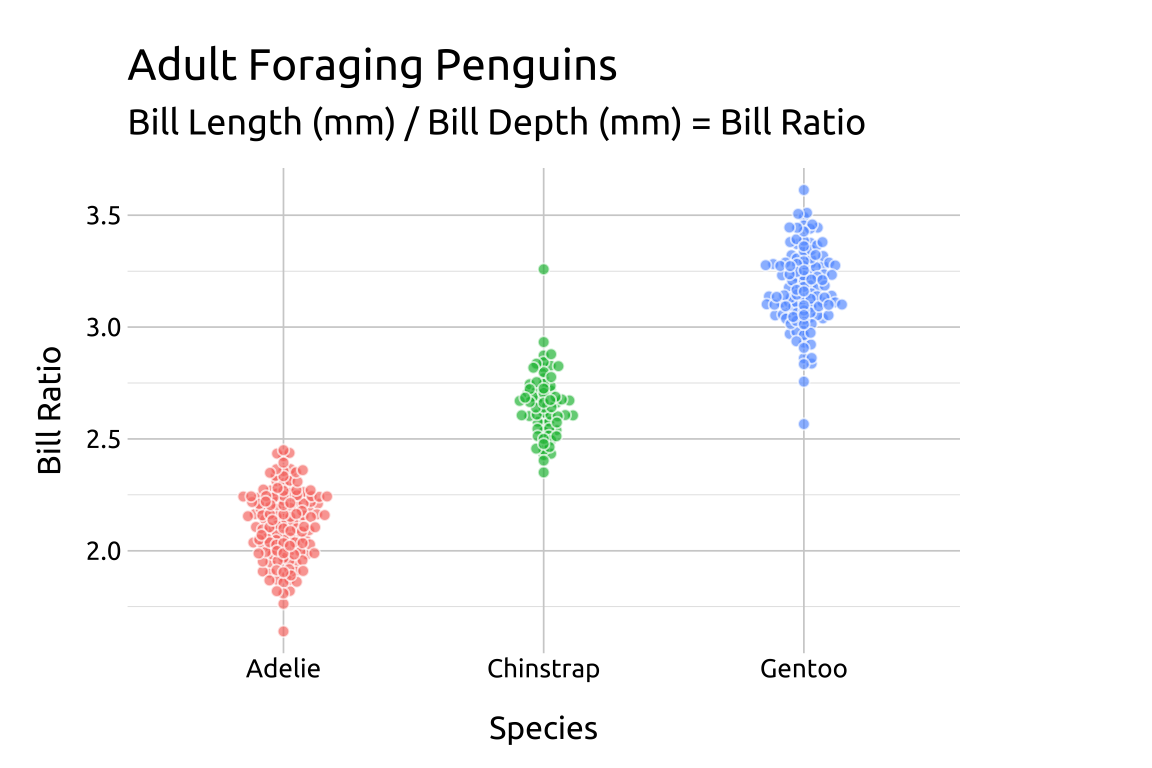
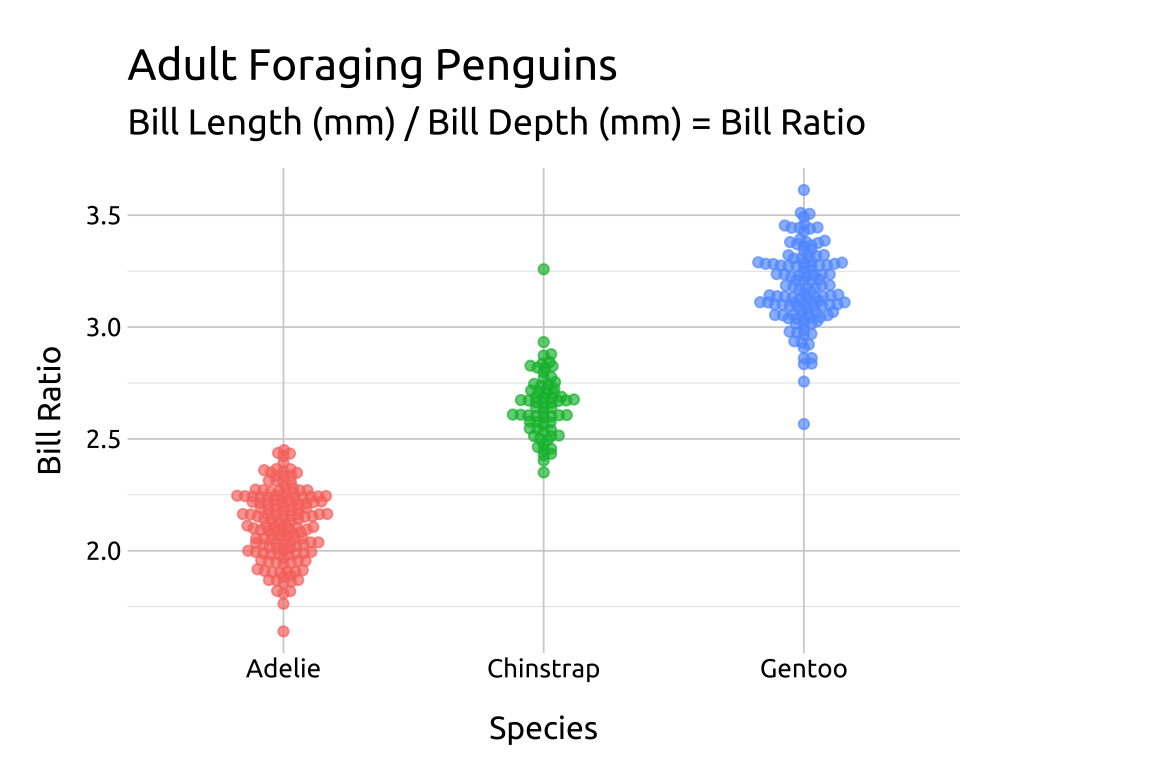
22.4.2 side
For a beeswarm that falls across the vertical axis, use the side argument.
show/hide
ggp2_rside_swarm <- ggplot(data = peng_beeswarm,
mapping = aes(x = species,
y = bill_ratio,
color = species)) +
ggbeeswarm::geom_beeswarm(
aes(fill = species),
side = 1, # right/upwards
shape = 21,
color = "#ffffff",
alpha = 2/3,
size = 1.7,
show.legend = FALSE)
ggp2_rside_swarm +
# add labels
labs_beeswarm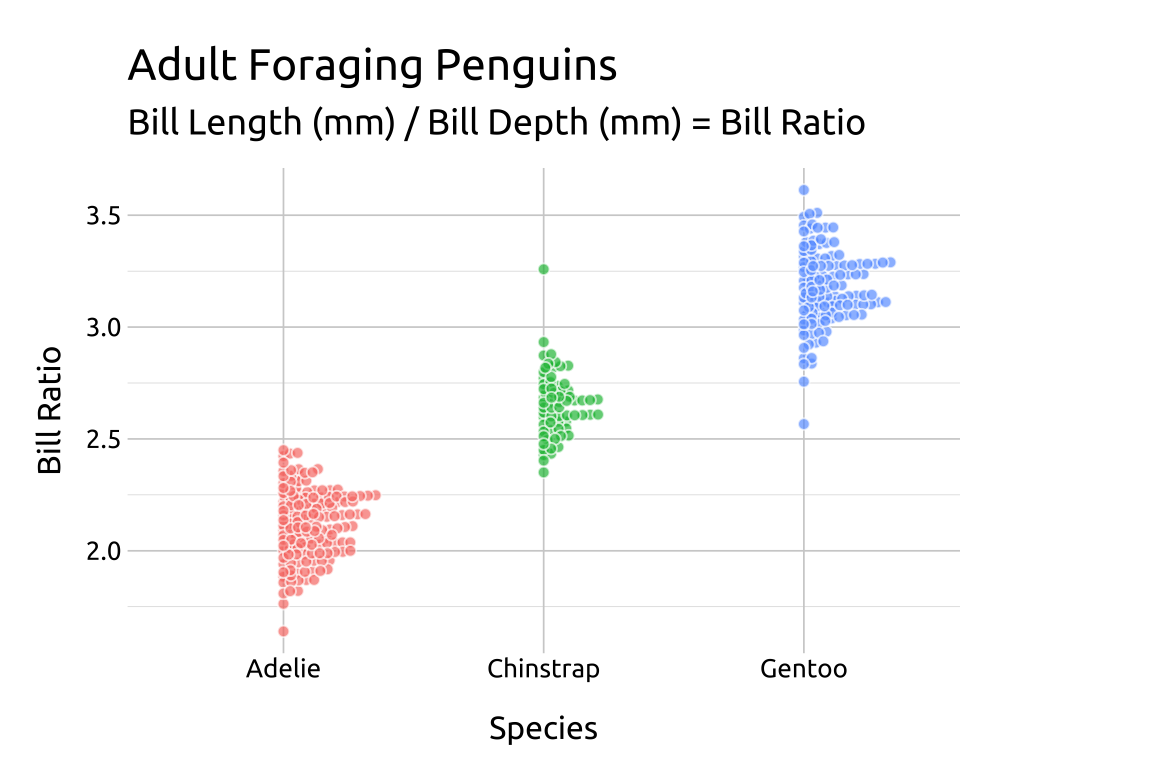
22.4.3 cex
The cex argument controls the “scaling for adjusting point spacing”
show/hide
ggp2_beeswarm_cex <- ggplot(data = peng_beeswarm,
mapping = aes(x = species,
y = bill_ratio,
color = species)) +
ggbeeswarm::geom_beeswarm(
aes(fill = species),
cex = 1.6,
shape = 21,
color = "#ffffff",
alpha = 2/3,
size = 1.7,
show.legend = FALSE)
ggp2_beeswarm_cex +
# add labels
labs_beeswarm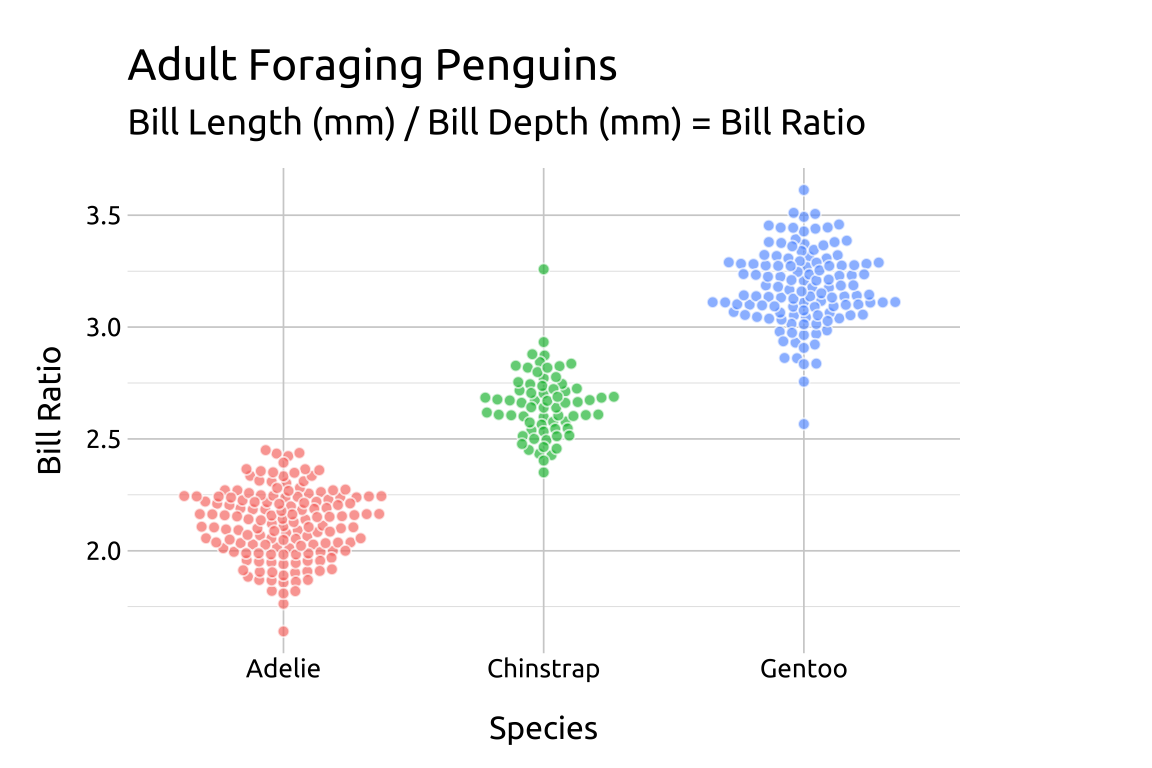
22.4.4 geom_jitter()
We can also create a beeswarm using the geom_jitter() and setting the height and width.
show/hide
ggp2_jitter_swarm <- ggplot(data = peng_beeswarm,
mapping = aes(x = species,
y = bill_ratio,
color = species)) +
geom_jitter(
aes(fill = species),
height = 0.05,
width = 0.11,
shape = 21,
color = "#ffffff",
alpha = 2 / 3,
size = 1.7,
show.legend = FALSE)
ggp2_jitter_swarm +
# add labels
labs_beeswarm