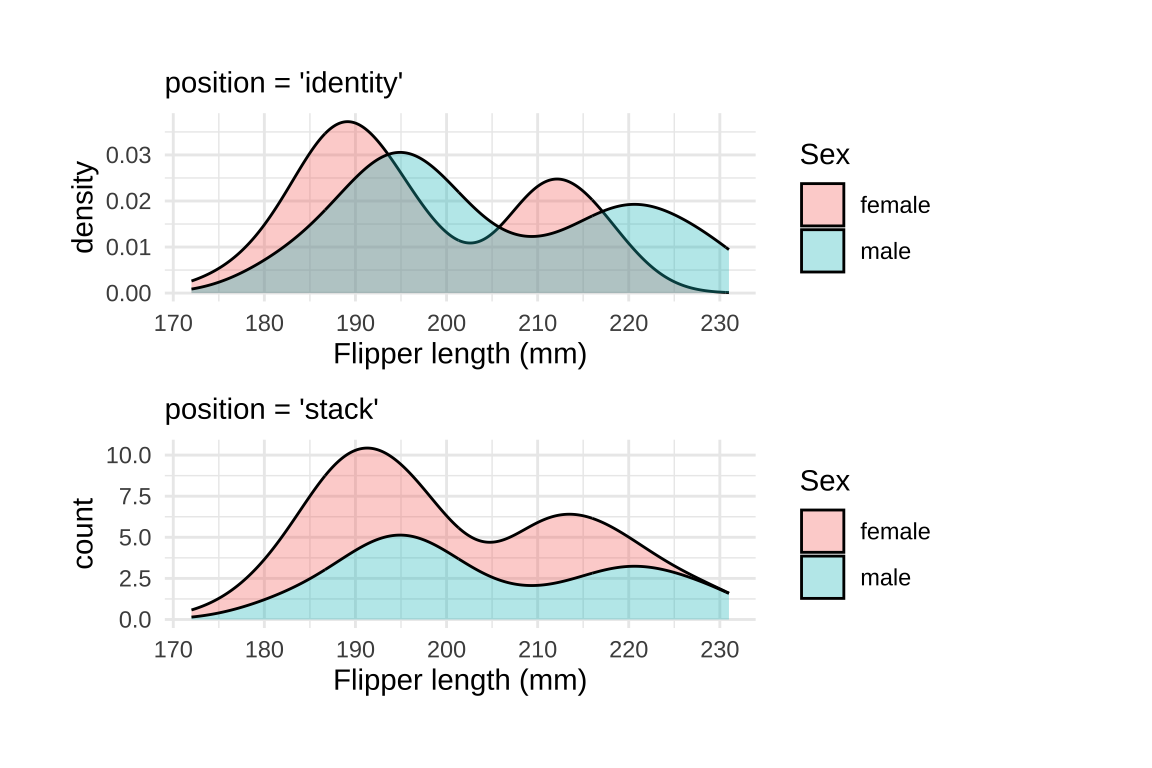
23 Overlapping density plot
23.1 Description
Density plots are smoothed version(s) of histogram(s). They can are great for comparing the distributions of a continuous variable across the levels of a categorical variable.
geom_density() creates a kernel density estimate. The default position argument is "identity", which takes the data as is. However, we can change position to "stack" to display overlapping distributions.
23.2 Set up
PACKAGES:
Install packages.
show/hide
install.packages("palmerpenguins")
library(palmerpenguins)
library(ggplot2)DATA:

Remove missing sex from the penguins data
show/hide
peng_density <- dplyr::filter(penguins, !is.na(sex))
dplyr::glimpse(peng_density)
#> Rows: 333
#> Columns: 8
#> $ species <fct> Adelie, Adelie, Adelie…
#> $ island <fct> Torgersen, Torgersen, …
#> $ bill_length_mm <dbl> 39.1, 39.5, 40.3, 36.7…
#> $ bill_depth_mm <dbl> 18.7, 17.4, 18.0, 19.3…
#> $ flipper_length_mm <int> 181, 186, 195, 193, 19…
#> $ body_mass_g <int> 3750, 3800, 3250, 3450…
#> $ sex <fct> male, female, female, …
#> $ year <int> 2007, 2007, 2007, 2007…23.3 Grammar
CODE:
Create labels with
labs()Initialize the graph with
ggplot()and providedataMap the
flipper_length_mmto thexandsextofillAdd the
geom_density()Set the
alphato1/3(to handle the overlapping areas)
show/hide
labs_ovrlp_density <- labs(
title = "Adult foraging penguins",
x = "Flipper length (millimeters)",
fill = "Sex")
ggp2_ovrlp_density <- ggplot(data = peng_density,
aes(x = flipper_length_mm,
fill = sex)) +
geom_density(alpha = 1/3)
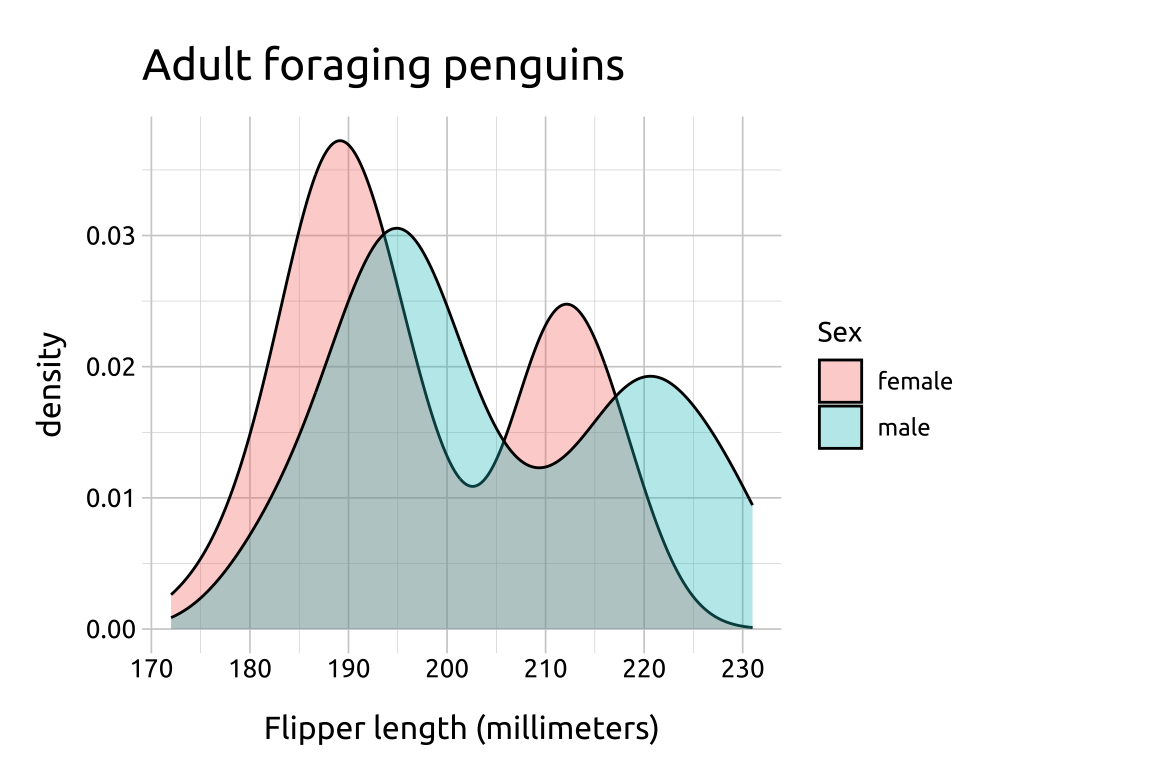
ggp2_ovrlp_density +
labs_ovrlp_densityGRAPH:
A downside of density plots is the lack of interpretability of the y axis
Make density area slightly transparent to handle over-plotting.
23.4 More info
ggplot2 has multiple options for overlapping density plots, so which one to use will depend on how you’d like to display the relative distributions in your data. We’ll cover three options below:
23.4.1 "stack"
If we change the position to "stack" we can see the smoothed estimates are ‘stacked’ on top each other (and the y axis shifts slightly).
show/hide
labs_stack_density <- labs(
title = "Adult foraging penguins",
x = "Flipper length (millimeters)",
fill = "Sex")
ggp2_stack_density <- ggplot(data = peng_density,
mapping = aes(x = flipper_length_mm,
fill = sex)) +
geom_density(position = "stack",
alpha = 1 / 3)
ggp2_stack_density +
labs_stack_density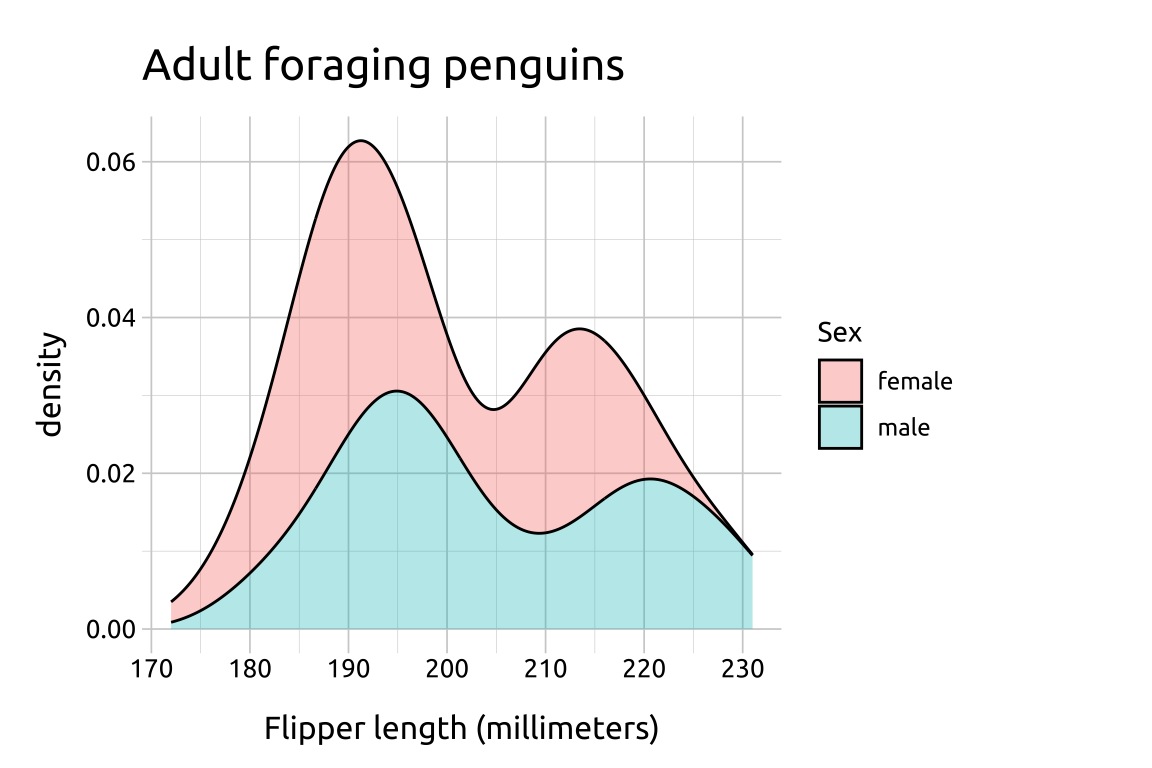
Setting position to 'stack' loses marginal densities
23.4.2 after_stat(count)
If we include after_stat(count) as one of our mapped aesthetics, the mapping is postponed until after statistical transformation, and uses the density * n instead of the default density.
show/hide
labs_after_stat_density <- labs(
title = "Adult foraging penguins",
x = "Flipper length (millimeters)",
fill = "Sex")
ggp2_after_stat_density <- ggplot(data = peng_density,
aes(x = flipper_length_mm,
after_stat(count),
fill = sex)) +
geom_density(position = "stack",
alpha = 1/3)
ggp2_after_stat_density +
labs_after_stat_density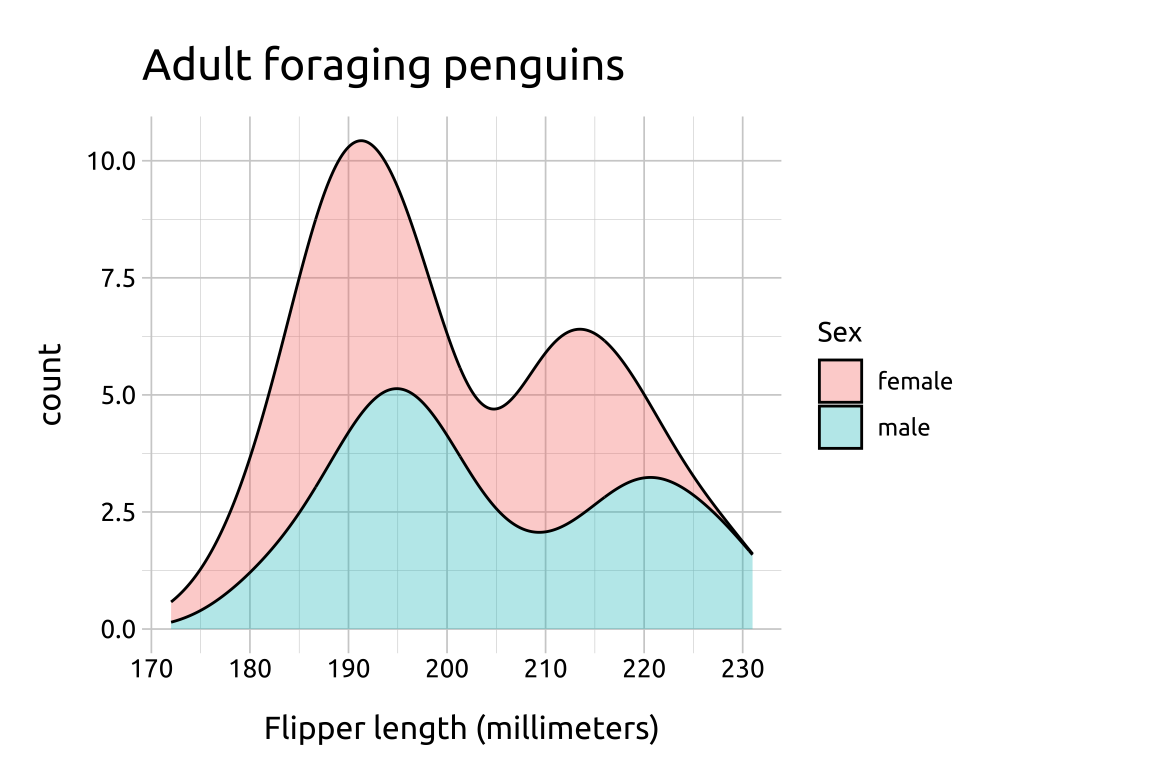
Adding after_stat(count) ‘preserves marginal densities.’, which result in more a interpretable y axis (depending on the audience)
23.4.3 "fill"
Using after_stat(count) with position = "fill" creates in a conditional density estimate.
show/hide
labs_fill_density <- labs(
title = "Adult foraging penguins",
x = "Flipper length (millimeters)",
fill = "Sex")
ggp2_fill_density <- ggplot(data = peng_density,
aes(x = flipper_length_mm,
after_stat(count),
fill = sex)) +
geom_density(position = "fill",
alpha = 1/3)
ggp2_fill_density +
labs_fill_density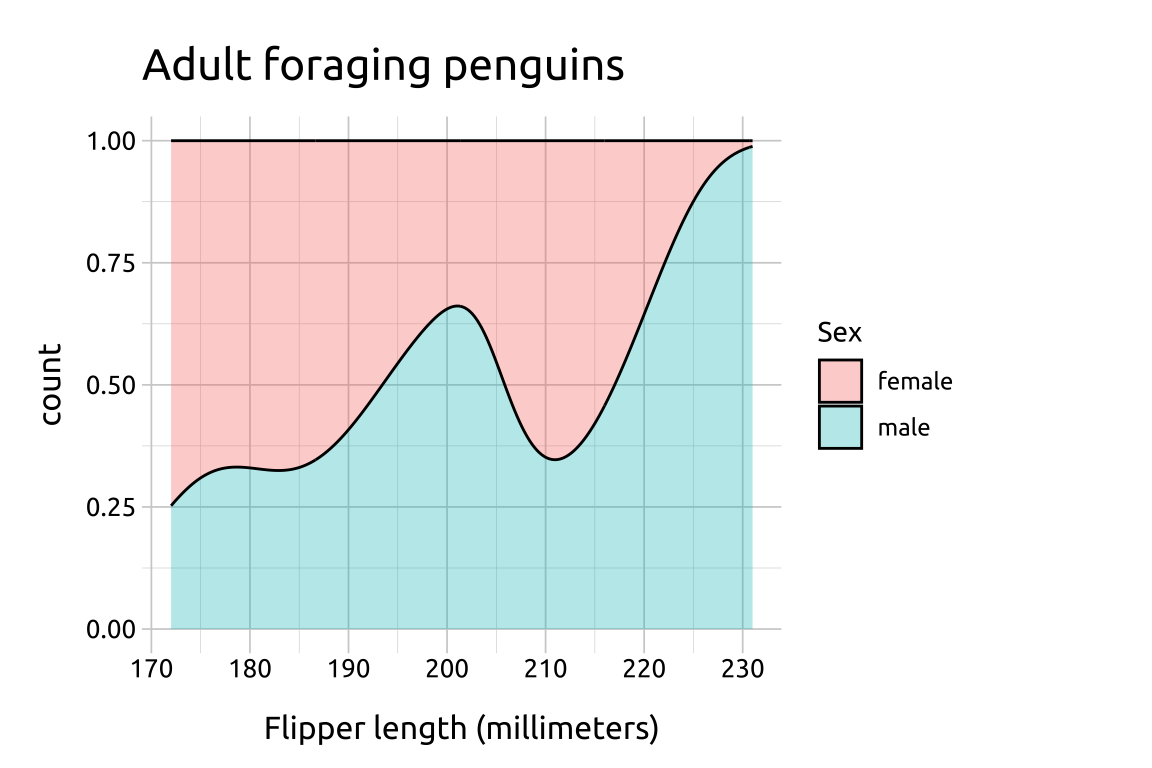
This results in a y axis ranging from 0-1, and the area filled with the relative proportional values.